Claudins are the main sealing protein of the intercellular tight junctions and play an important role in cancer progression and dissemination. Several authors have reported conflicting results of claudin-1 expression in colorectal carcinoma. The goal of this study is to examine claudin-1 expression in a tissue microarray of colorectal cancer and metastases, simultaneously, in order to assess the prognostic value of this protein in colorectal cancer. This study analysed the expression of claudin-1 by immunohistochemistry in 99 tissue samples (including 33 colorectal cancer cases with 33 concordant lymph node metastases and 33 concordant cancer-adjacent normal tissues, all from the same patients). The tumours included adenocarcinomas from the colon (n=25) and from the rectum (n=8). This is the first study to demonstrate the expression of claudin-1 protein across a sample of colorectal cancer and its metastases, simultaneously. We report loss of claudin-1 expression in the lymph node metastases in the majority of cases. We also report differential expression of claudin-1 among colorectal cancers of different grades, with the highest expression being in well-differentiated, low-grade tumours; and complete loss of expression in the majority of cases of poorly-differentiated, high grade tumours.
colon, cancer, claudin-1, tight junction proteins, adenocarcinoma
Claudins are tight junction proteins which, along with adherens junctions and desmosomes form cellular sheets. Tight junctions (TJs) are critical for sealing of cellular sheets, thereby controlling paracellular ion flux. In addition to that, tight junctions also play critical roles in maintaining cell polarity and signal transductions [1,2]. Claudins are the backbone of tight junction strands, and are essential components to the function of tight junctions [1].
The high degree of cellular organization typically observed in normally differentiated tissues is often lost in cancer. Loss of epithelial integrity with changing claudins’ levels and resultant increased para-cellular leakage plays a critical role in providing a space for tumor cell mobility and increased nutrients’ supply for tumor cells underneath.
Several studies analyzing the roles of tight junctions in oncogenesis have revealed either downregulation or upregulations of claudins' expression [3-10]. The downregulation and loss of these proteins are expected events in carcinogenesis. As the cell becomes malignant, it loses attachment to underlying basement membrane and nearby cells during which the TJs are disrupted and lost. It is interesting to note, though, that numerous other studies have shown overexpression of TJs in cancer cells of different types. It is likely those overexpression events are related to roles these genes play other than paracellular seal formation [11]. It is through those “other” functions [1,2] that TJs and especially claudins play a role in carcinogenesis. In conclusion, claudins, whether upregulated or downregulated, are proving to be a vital piece of the puzzle of oncogenesis.
A prime example of the aforementioned role of claudins in carcinogenesis and metastases is claudin-1. It is one of the genes strongly regulated by B-catenin [12]. The latter gene is known for its role in maintaining cell-to-cell adhesion and most importantly in mediating the oncogenic Wnt/B-catenin transduction pathway. Claudin-1 along with claudin-3 and claudin-5 are shown to promote pro-MMP2 whereby claudins recruit MMPs on the cell surface to achieve elevated focal concentrations and eventual activations of pro-MMP2 [12].
Claudin-1 expression is frequently altered in several cancers, including upregulation and down regulation [13]. In Colorectal cancer, claudin-1 plays an important role in the oncogenesis and has been proven to have a prognostic value [14]. Dhawan et al. [15] showed that claudin-1 upregulation contributes to an aggressive and metastatic phenotype, while its knock-down lead to reduction in the B-catenin/Tcf/Lef signaling [15].
This and other studies have confirmed the role of claudin-1 upregulation in colon carcinogenesis and metastases. However, regrettably, conflicting and contradictory results have been reported by several authors regarding claudin-1 expression in colorectal cancer. Some authors are reporting suppressed claudin-1 expression in colorectal cancer and linked this with depth of invasion, histological grade and lymph-vascular/perineural invasion [16-21]. Resnick et al. [21] went further to affirm that loss of claudin-1 expression is a strong predictor of disease recurrence, and poor patient survival in stage II colon cancer. Other studies on the other hand, have shown increased expression of claudin- 1 in CRC cells [12,15,22-24] resulting in increased depth of tumor invasion [15,23] through activation of the matrix metalloproteinases [25].
This contradiction has dealt a blow against using claudin-1 as a biomarker of prognosis in colorectal carcinoma. Without reliable results, a protein biomarker can never be used as such. This is the first comprehensive study analysing claudin-1 expression in colorectal cancer (CRC) and its lymph node metastases simultaneously. By doing this, we hope to paint a complete picture of the changes seen in claudin-1 expression in normal, cancerous and metastatic sample in the same patient. In the process of doing so, we try to find some answers for the apparent contradiction above and to also give back some credence to the claudin-1 expression in colorectal cancer.
Colorectal carcinoma tissues
Using commercial human colon carcinoma cells’ tissue microarray (Catalog no. colon991, Biomax, US, Rockville, Maryland), 99 tissue samples/cores (including 33 colorectal cancer cases with 33 concordant lymph node metastases and 33 concordant cancer-adjacent normal tissues, single core per case) were analyzed for claudin-1 expression by immunohistochemistry. The tumors included adeno-carcinomas from the colon (n=25) and from the rectum (n=8). The samples from which our tissue array was derived are usually put in formalin within 15 minutes following surgical resection. Two tissue sections of every lot of tissue arrays are sampled for immunohistochemistry studies to ensure the validation of antigenicity remains in the tissue. Our sample consists of cores ranging from 1.0 to 1.5 mm in diameter.
Immunohistochemistry
A rabbit polyclonal antibody against human claudin-1 (RB-9209-P, Labvision, Fremont, California) was diluted 1:300 in antibody diluent (Dako Co., Mississauga, ON, Canada) and applied to 5-um-thick sections from formalin-fixed, paraffin-embedded tissue specimens, using the avidin-biotin peroxidase method (Vectastatin Elite ABC kit, Vector Laboratories, Burlingame, California), following the manufacturer’s instructions. The immunohistochemical (IHC) stain was performed manually at room temperature. Negative controls were used with omission of primary antibody. Separate positive controls of normal skin were used for test optimization and run validation.
Scoring and analysis of positive immunostaining
Claudin-1 staining intensity was evaluated semiquantitatively and graded as follows: Weak (absent or weakly present, 0 to 1) = staining intensity less than normal skin, Moderate (2) = staining similar to normal skin and Strong (3) = Staining intensity higher than normal skin.
All tissues, normal controls, tissues adjacent to cancer or malignant tissue, were compared to the staining of normal skin. Only cellular membrane immunoreactivity was used in the evaluation. While cytoplasmic and nuclear claudin-1 expressions were documented, it did not connote any differential treatment merely by its presence. The immunostaining for quantity was assessed as follows: 0, 0 cells positive, 1, less than 25% of cells are positive, 2, 25% to 50% of cells positive, 3, 50% to 75% of cells positive, 4, greater than 75% of cells positive. All assessments were done by two investigators (A.O. and RN) independently and then jointly to agree at the final scoring.
The patients had a median age of 58 years (range from 20 to 79). The group comprised of 10 females and 23 males. Tumours were graded as follows: grade I (2 cases), grade II (21 cases), grade III (7 cases) and grade IV (3 cases). Cores of cancer-adjacent normal tissue were included. The IHC stain result was not reported in one of the lymph node metastasis of a colorectal carcinoma (D-2) and in a cancer adjacent normal appearing tissue of a rectal carcinoma (J-3), because tissues were missing in both cases.
Claudin-1 expression was primarily membranous in the majority of well-differentiated colorectal carcinoma (CRC) cases of our study (Figure 1A), although faint to strong cytoplasmic and nuclear staining were also seen in some of these cases (Figure 1B). Overall, 16/33 colorectal carcinoma cases expressed claudin-1, among them were 13 grade II colorectal carcinomas (13/21), one grade I colorectal carcinoma (1/2), two grade III (2/7) colorectal carcinomas and zero (0/3) grade IV CRC. Poorly differentiated CRCs (grades III and IV) were mostly negative (Figure 1C). Four (4) lymph nodes of 33 lymph nodes in total expressed claudin-1 expression (Figure 1D). All those 4 lymph nodes were metastases from tumours which expressed claudin-1. The 4 lymph nodes were divided as follows: There was 1 claudin1-positive lymph node of grade II metastatic CRC, 2 claudin1-positive lymph nodes’ of grade III metastatic CRC and 1 claudin1-positive lymph node of grade I metastatic CRC. None (0/33) of the cancer-adjacent normal tissue expressed claudin-1 (Figure 1E). While the results were not statistically significant, possibly due to small sample size, it is worth further investigation. Detailed staining results of the colorectal carcinoma and metastases cases are summarized in Table1.
Table 1. The numbers in red in the parentheses are the grades of tumours
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
A |
0 (I) |
0 |
0 |
2x3 (1) |
2x3 |
0 |
1x1 (II) |
0 |
0 |
B |
0 (II) |
0 |
0 |
1x2 (II) |
0 |
0 |
0 (II) |
0 |
0 |
C |
1x1 (II) |
0 |
0 |
1x3(II) |
0 |
0 |
1x1 (II) |
0 |
0 |
D |
1x1 (II) |
Missing |
0 |
0 (II) |
0 |
0 |
0 (II) |
0 |
0 |
E |
1x1 (II) |
0 |
0 |
0 (II) |
0 |
0 |
0 (IV) |
0 |
0 |
F |
1x1 (II) |
0 |
0 |
0 (II) |
0 |
0 |
1x1 (II) |
2x4 |
0 |
G |
1x1 (II) |
0 |
0 |
1x1 (II) |
0 |
0 |
1x1 (II) |
0 |
0 |
H |
1x1 (II) |
0 |
0 |
0 (III) |
0 |
0 |
0 (II) |
0 |
0 |
I |
0 (III) |
0 |
0 |
0 (II) |
0 |
0 |
0 (III) |
0 |
0 |
J |
0 (IV) |
0 |
Missing |
0 (III) |
0 |
0 |
0 (IV) |
0 |
0 |
K |
1x1 (III) |
1x3 |
0 |
2x1 (III) |
1x3 |
0 |
0 (III) |
0 |
0 |
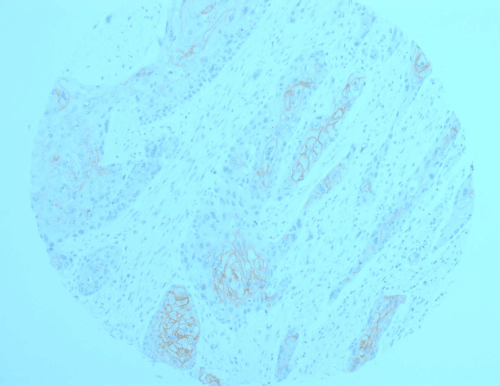
Figure 1A. Representative section of colorectal carcinoma (CRC) where well-differentiated areas express membranous pattern of staining of claudin-1 protein, while poorly differentiated areas are negative for this protein
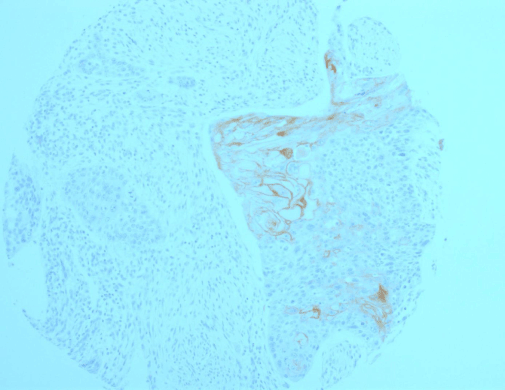
Figure 1B. A CRC exhibiting heterogeneous morphology with well-differentiated areas highlighted by a brisk claudin-1 expression, and poorly-differentiated areas with no biomarker expression. Also noted are the focal, strong cytoplasmic and nuclear claudin-1 expressions in addition to the membranous one in well-differentiated areas of the tumour.
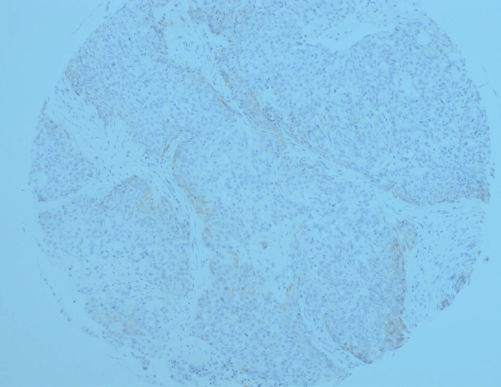
Figure 1C. Absence of claudin-1 expression in poorly-differentiated CRC.
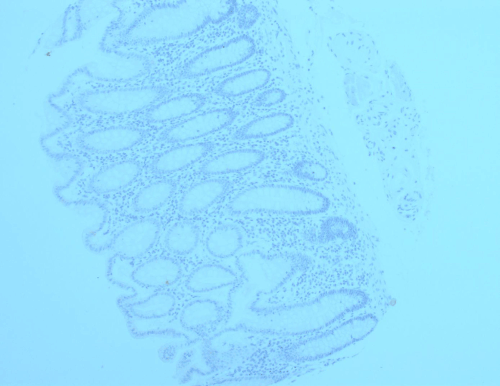
Figure 1D. A representative section of cancer-adjacent, normal-appearing colonic mucosa exhibiting no claudin-1 expression.
The numbers in black, are the IHC intensity score (first number) graded from 0 to 3, and the second number is the quantity of positive cells graded from 0 to 4.
Tumor grade and staining score
It is clear in our study that there a link between staining score and tumour grade, with well- to moderately-differentiated tumours/regions (grades I and II) expressing strong staining (Figures 1A and 1B), while the poorly-differentiated tumours (grades III and IV) were negative (Figure 1C). Some tumours showed heterogeneous morphology with mixed areas of grades I/II and grades III/IV. It was interesting to note that areas of better differentiation expressing the marker, while least differentiated areas went silent (Figure 1A and 1B).
Furthermore, tumours showing the highest expression (grades I and II) were also showing focal, faint to strong cytoplasmic and nuclear claudin-1 staining, in addition to the membranous staining found (Figure 1B).
Normal tissues
Staining of sections taken from cancer-adjacent, normal- appearing colon or rectal mucosa is also summarized in Table 1. Virtually all sections taken from cancer-adjacent normal colorectal mucosa expressed no staining at all (0, absent staining, less than normal skin, Figure 1D). Other studies have shown similar results: Grone et al. [24] reported only 4/16 normal colonic mucosa cases expressing very weak claudin-1 expression. The rest of normal colonic mucosa sections (12 sections) were negative for claudin-1. Dhawan et al. and Miwa et al. [12,15] reported no claudin-1 expression in normal colonic mucosa taken from colorectal cancer patients.
This is the first study to comprehensively report and analyze the expression of claudin-1 in CRCs and its lymph node metastases, simultaneously. In line with other studies [12,15,22,24] , ours does show that claudin-1 expression is increased in colorectal cancer compared to normal colonic mucosa. This overexpression is seen in 50% of case (16/33). We are the first to report that the majority of colorectal metastases lose its expression of claudin-1 [12]. CRC lymph node metastases lost its claudin-1 expression from a total of 16 metastases, 75%). Thirdly, it is also apparent that claudin-1 is more likely to be overexpressed in low grade, well-differentiated tumors, with sixty-two percent (62%) of grade II CRCs overexpressing claudin-1, 29% of grade III CRCs overexpressing claudin-1, and 0% expression in grade IV CRCs. No expression of claudin-1 is seen in any of the CRC metastases which has a parent- tumor negative for claudin-1.
The role of claudin-1 in CRC development and progression could be summarized as follows. The apparent overexpression of claudin-1 results in increased permeability and leakiness of the paracellular spaces and significant disorganization of the TJ structure [26]. This was evident by the increased paracellular permeability to ruthenium red [26]. Increased permeability of TJs is a tumor-promoting event in epithelial cancer [27, 28]. Secondly, Claudin-1 is one of the genes strongly related to B-catenin which is known for mediating the oncogenic Wnt/beta-catenin transduction pathway [12]. And thirdly, claudin-1, along with claudins-3 and -5, are shown to promote pro-MMP2, whereby claudins recruit MMPs on the cell surface to achieve elevated focal concentrations and eventual activation of pro_MMP2 [13].
One reason why the expression of claudin-1 turned negative or lost by protein studies but not by genomic expression analyses’ tools; maybe that the sensitivity of genomic expression analyses’ tools far outweighs those of protein analyses. Another reason for the above is that earlier studies have relied on bulk sampling, evident by the fact that many of these studies have reported heterogeneous staining pattern of claudin-1 in their samples [29]. This methodology of using bulk sampling, which usually includes normal benign as well as tumorous cells, will tend to obscure changes of mRNA and protein expressions in tumour and normal epithelium [24]. Since laser micro-dissection enables selective dissection of cells of interest; accurate expression analyses of groups of genes and proteins are now feasible focusing biopsies on viable tissue. Our samples were taken from viable, non-necrotic regions as evident by histologic assessment. On the other hand, other authors believe that the depressed claudin-1 levels they observed were the result of using IHC or Western blot and not mRNA expression studies [16]. In our opinion, this may be possible if the actual half-life of claudin-1 proved to be short. It is known that discrepancy between results of Quantitative Real Time (QRT) Polymerase Chain Reaction (PCR) and proteins studies may well happen due to short half-life of the protein. So here we would like to emphasize the need for further studies exploring the half-life of claudin-1 protein.
2021 Copyright OAT. All rights reserv
While we agree that loss of claudins and resultant increased cell motility and invasion have a role to play in carcinogenesis [16,30]; we also believe that this loss of expression is preceded by an earlier event of overexpression of the proteins in question. Studies have reported that colonic adenoma cells have increased expression of claudin-1 [31]. This, combined with results showing increased expression of claudin-1 in grades I and II and loss in more advanced grades [8,15,24 and this study], and with our findings in this study of loss of claudin-1 expression in metastatic lymph node lesions, all of this suggest that overexpression of this protein is an early event in CRC development, necessary for the next step, i.e., invasion of underlying tissues through the activation of the matrix metalloproteinases (MMPs) system. Once that invasion is complete, claudin-1 expression is lost, as is evident in our study in the lymph node metastatic lesions. Our findings in this study are similar to previous work done on oral squamous cell carcinoma [32], where it became clear that claudin-1 expression is a “power-up” mechanism, where a squamous cell carcinoma in-situ cell experiences increased expression of claudin-1, followed by invasion of the basement membrane. The expression of claudin-1 will be lost once invasion is complete. This was also the case with metastatic melanoma. Cohn et al. [33] reported strong correlation between claudin-1 expression in primary melanoma and depth of Clark level/Breslow invasion, in contrast to metastatic melanoma which loses claudin-1 expression in the vast majority of cases. The same happens in cervical neoplasia. Claudin-1 expression gradually increases in accordance with the progression of the lesions from normal to low-grade squamous intraepithelial lesion to high-grade intra-epithelial lesion, and then it becomes lost in invasive carcinoma [34,35].
In light of the above, claudin-1 over-expression in colorectal cancer appears to show up as a prelude to tumor invasion, and once invasion is complete, the tumor cell would lose this expression in the majority of cases. As a result of the above, we hereby recommend that the use of claudin-1 staining include carcinoma-in-situ (CIS) lesions and dysplastic adenomatous polyps as well as invasive/metastatic lesions in the colon.
In conclusion, in this study we have shown overexpression of claudin-1 protein in approximately 50% of colorectal cancers, with loss of claudin-1 expression in lymph nodes’ metastatic CRC lesions in 75% of cases, and a differential expression of claudin-1 between well-differentiated and poorly-differentiated tumours. This over-expression and eventual loss of claudin-1, as seen in poorly-differentiated and metastatic tumours, may be useful in diagnostic and prognostic modalities of CRC. Our understanding of the exact function and role of claudin-1 in epithelial tumorigenesis is increasing, and there is significant potential in applying our research findings in the management of neoplastic conditions, both diagnostically and prognostically.
- Tsukita S, Furuse M (2000) Pores in the wall: claudins constitute tight junction strands containing aqueous pores. J Cell Biol 149: 13-16. [Crossref]
- Van Itallie CM, Rogan S, Yu A, Vidal LS, Holmes J, et al. (2006) Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am J Physiol Renal Physiol 291: F1288-1299. [Crossref]
- Krämer F, White K, Kubbies M, Swisshelm K, Weber BH (2000) Genomic organization of claudin-1 and its assessment in hereditary and sporadic breast cancer. Hum Genet 107: 249-256. [Crossref]
- A Moustafa AE, Alaoui-Jamali MA, Batist G, Hernandez-Perez M, Serruya C, et al. (2002) Identification of genes associated with head and neck carcinogenesis by cDNA microarray comparison between matched primary normal epithelial and squamous carcinoma cells. Oncogene 21: 2634-40. [Crossref]
- Kominsky SL (2006) Claudins: emerging targets for cancer therapy. Expert Rev Mol Med 8: 1-11. [Crossref]
- Long H, Crean CD, Lee WH, Cummings OW, Gabig TG (2001) Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res 61: 7878-7881. [Crossref]
- Rangel LB, Agarwal R, D'Souza T, Pizer ES, Alò PL, et al. (2003) Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res 9: 2567-7.
- Dos Reis PP, Bharadwaj RR, Machado J, Macmillan C, Pintilie M, et al. (2008) Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer 113(11):3169-80. [Crossref]
- Oku N, Sasabe E, Ueta E, Yamamoto T, Osaki T (2006) Tight junction protein claudin-1 enhances the invasive activity of oral squamous cell carcinoma cells by promoting cleavage of laminin- 5 gamma2 chain via matrix metalloproteinase (MMP)-2 and membrane-type MMP-1. Cancer Res 66: 5251-7. [Crossref]
- Ouban A, Hamdan H, Hakam A, Ahmed AA (2012) Claudin-1 expression in squamous cell carcinomas of different organs: comparative study of cancerous tissues and normal controls. Int J Surg Pathol 20: 132-138. [Crossref]
- Hewitt KJ, Agarwal R, Morin PJ (2006) The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer 6: 186. [Crossref]
- Miwa N, Furuse M, Tsukita S, Niikawa N, Nakamura Y, et al. (2001) Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res 12: 469-476. [Crossref]
- Miyamori H, Takino T, Kobayashi Y, Tokai H, Itoh Y, et al. (2001) Claudin promotes activation of pro-matrix metalloproteinase-2 mediated by membrane-type matrix metalloproteinases. J Biol Chem 276: 28204-28211. [Crossref]
- Ouban A, Ahmed AA (2010) Claudins in human cancer: a review. Histol Histopathol 25: 83-90. [Crossref]
- Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, et al. (2005) Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest 115: 1765-1776. [Crossref]
- SÃren D, YÄldÄrÄm M, Kaya V, AlikanoÄŸlu AS, BÃlbÃller N, et al. (2014) Loss of tight junction proteins (Claudin 1, 4, and 7) correlates with aggressive behavior in colorectal carcinoma. Med Sci Monit 20: 1255-1262. [Crossref]
- Shibutani M, Noda E, Maeda K, Nagahara H, Ohtani H, et al. (2013) Low expression of claudin-1 and presence of poorly-differentiated tumor clusters correlate with poor prognosis in colorectal cancer. Anticancer Res 33: 3301-3306. [Crossref]
- Abdelzaher E, Rizk AM, Bessa SS, Omer KM (2011) Predictive value of immunohistochemical expression of claudin-1 in colonic carcinoma. J Egypt Natl Canc Inst 23: 123-131. [Crossref]
- Yoshida T, Kinugasa T, Akagi Y, Kawahara A, Romeo K, et al. (2011) Decreased expression of claudin-1 in rectal cancer: a factor for recurrence and poor prognosis. Anticancer Res 31: 2517-2525. [Crossref]
- Nakagawa S, Miyoshi N, Ishii H, Mimori K, Tanaka F, et al. (2011) Expression of CLDN1 in colorectal cancer: a novel marker for prognosis. Int J Oncol 39: 791-796. [Crossref]
- Resnick MB, Konkin T, Routhier J, Sabo E, Pricolo VE (2005) Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol 18: 511-518. [Crossref]
- Tang W, Dou T, Zhong M, Wu Z (2011) Dysregulation of Claudin family genes in colorectal cancer in a Chinese population. Biofactors 37: 65-73. [Crossref]
- Huo Q, Kinugasa T, Wang L, Huang J, Zhao J, et al. (2009) Claudin-1 protein is a major factor involved in the tumorigenesis of colorectal cancer. Anticancer Res 29: 851-857. [Crossref]
- Gröne J, Weber B, Staub E, Heinze M, Klaman I, et al. (2007) Differential expression of genes encoding tight junction proteins in colorectal cancer: frequent dysregulation of Claudin-1, -8 and -12. Int J Colorectal Dis 22: 651–59. [Crossref]
- Takehara M, Nishimura T, Mima S, Hoshino T, Mizushima T (2009) Effect of claudin expression on paracellular permeability, migration and invasion of colonic cancer cells. Biol Pharm Bull 32: 825-831. [Crossref]
- de Oliveira SS, de Oliveira IM, De Souza W, Morgado-Díaz JA (2005) Claudins upregulation in human colorectal cancer. FEBS Lett 579: 6179-6185. [Crossref]
- Weinstein RS, Merk FB, Alroy J (1976) The structure and function of intercellular junctions in cancer. Adv Cancer Res 23: 23-89. [Crossref]
- Polak-Charcon S, Shoham J, Ben-Shaul Y (1980) Tight junctions in epithelial cells of human fetal hindgut, normal colon, and colon adenocarcinoma. J Natl Cancer Inst 65: 53-62. [Crossref]
- Kinugasa T, Huo Q, Higashi D, Shibaguchi H, Kuroki M, et al. (2007) Selective up-regulation of claudin-1 and claudin-2 in colorectal cancer. Anticancer Res 27: 3729-3734. [Crossref]
- Singh AB, Sharma A, Dhawan P (2010) Claudin family of proteins and cancer: an overview. J Oncol 2010: 541957. [Crossref]
- Bezdekova M, Brychtova S, Sedlakova E, Langova K, Brychta T, et al. (2012) Analysis of Snail-1, E-cadherin and claudin-1 expression in colorectal adenomas and carcinomas. Int J Mol Sci 13: 1632-1643. [Crossref]
- Ouban A, Ahmed A (2014) Analysis of the Distribution and Expression of Claudin-1 Tight Junction Protein in the Oral Cavity. Appl Immunohistochem Mol Morphol. [Crossref]
- Cohn ML, Concharuk VN, Diwan AH, Zhang PS, Shen SS, et al. (2005) Loss of claudin-1 expression in tumor- associated vessels correlates with acquisition of metastatic phenotype in melanocytic neoplasms. J Cutan Pathol 32: 533-536.[Crossref]
- Lee JW, Lee SJ, Seo J, Song SY, Ahn G, et al. (2005) Increased expressions of claudin-1 and claudin-7 during the progression of cervical neoplasia. Gynecol Oncol 97: 53-59. [Crossref]
- Sobel G, Paska C, Szabo I, Kiss A, Kádár A, et al. (2005) Increased expression of claudins in cervical squamous intraepithelial neoplasia and invasive carcinoma. Hum Pathol 36: 162-169. [Crossref]




