Background: To understand pathophysiology of systemic lupus erythematosus (SLE), we here tried to identify profiles of cell surface (CS) molecules of peripheral blood mononuclear cells (PBMCs) using targeted proteomics.
Methods: PBMCs prepared from 5 patients with SLE and 5 healthy donors (HLs), were subjected to live cell-biotinylation. Then the biotinylated CS proteins were analyzed by 2-dimensional fluorescence difference gel electrophoresis (2D-DIGE). Protein spots that showed different intensity between the SLE and HL groups were identified by mass spectrometry.
Results: In total, 468 protein spots were detected by 2D-DIGE, out of which 151 spots (32.3%) showed significantly different intensity between the two groups. Among the 151 spots, 137 spots (29.3%) showed > ±1.5-fold different intensity and 44 spots (9.4%) showed > ±2.5-fold different intensity between the two groups. Proteins in 17 out of the 44 spots were identified.
Conclusion: Our study comprehensively investigated CS protein profiles of PBMCs in SLE for the first time, to our knowledge. We found that CS protein profiles of PBMCs from patients with SLE were greatly different from those from HLs. Our study would provide a new strategy to investigate the pathophysiology of SLE.
surface proteins, systemic lupus erythematosus, proteomics
Systemic lupus erythematosus (SLE) is a prototype of systemic autoimmune diseases, characterized by generation of various autoantibodies. Patients with SLE display clinical features in multiple organs such as kidney, skin, and central nerve systems and also display various abnormalities in laboratory examinations like anti-nuclear antibodies (ANAs) and low levels of serum complements. Since the clinical features and abnormalities are considerably different among patients with SLE, the diagnosis of SLE is made by referring the revised classification criteria for SLE 1997 [1] or more recently by the systemic lupus international collaborating clinics classification criteria for SLE 2012 [2,3]. Only a limited part of autoantibodies such as SS-A antibodies has been evidenced to cause damages of an organ [4,5]. However, direct roles of the remaining large part of the autoantibodies in the clinical features have not been elucidated.
Although mechanisms for the autoantibody production in SLE have not been fully understood, both B cells and T cells are thought to be involved in the autoantibody production [6,7]. For example, expansion of memory B cells has been demonstrated and thus SLE is suggested to be characterized by frequent cycles of B cell activation and differentiation [8]. T cells are thought to abnormally assist the activation and differentiation of autoreactive B cells. Expansion of the Th17 population and perturbation of T cell receptor (TCR) physiology and downstream signaling of TCRs have been reported [9]. However, molecular mechanisms for the alterations in T and B cells have been not fully understood.
In general, any type of cells possesses various molecules on their surface. Cell surface (CS) molecules that receive extracellular information mediate intracellular signal transduction. Thus, abnormal expression or post-translational modification of CS molecules can be directly linked to the pathophysiology of SLE. As a tool to analyze CS proteins, a fluorescence-activated cell sorting (FACS) analysis has been established. In fact, FACS has provided various findings on abnormalities of CS proteins of PBMCs in SLE [10-13]. However, FACS can analyze only limited kinds of CS proteins and thereby cannot comprehensively analyze CS proteins. Based on these backgrounds, we here tried to comprehensively analyze CS protein profiles using targeted proteomics, the combination of live cell-biotinylation and 2-dimensional fluorescence difference gel electrophoresis (2D-DIGE), to understand pathophysiology of SLE.
Clinical specimens
PBMCs were isolated by density-gradient centrifugation using Ficoll-Paque Plus (Pharmacia Biotech, Uppsala, Sweden) from blood samples of 5 patients with SLE and 5 healthy donors (HLs). All the samples were obtained with written informed consent and this study was approved by the institutional ethics committee of St. Marianna University School of Medicine.
Isolation of CS proteins by biotin labeling
CS proteins were isolated by a commercially available CS protein isolation kit (Thermo Fisher Scientific Inc., Waltham, MA, USA), according to the manufacturer's instructions. Briefly, CS proteins of live PBMCs were biotinylated with sulfo-NHS-S-S-biotin, which was a cell-impermeable and cleavable biotinylation reagent. After the biotinylation, PBMCs were lysed and the biotinylated CS proteins were recovered by avidin beads (Thermo Scientific NeutrAvidin Agarose Resin). Then CS proteins were released from the avidin-bound biotin by cleavage of the S-S bonds using a reducing reagent.
2-dimensional fluorescence difference gel electrophoresis (2D-DIGE)
The isolated CS proteins were separated by 2D-DIGE. 2D-DIGE was performed as described previously [14]. Each of the CS protein samples was labeled with Cyanine dye 5 (Cy5, Cy Dye DIGE Saturation dye; GE Healthcare, Buckinghamshire, UK). An internal control “standard sample” was prepared by mixing an equal amount of each of the samples. The standard sample was then labeled with Cyanine dye 3 (Cy3, GE Healthcare). Then, each of the Cy5-labeled protein samples (2.5 µg) were mixed with the Cy3-labeled standard sample (2.5 µg). The mixture was applied onto an isoelectric focusing (IEF) gel (pH 3-11, GE Healthcare). Subsequently, proteins separated by IEF were further separated by 12.5 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were scanned using an image analyzer (Typhoon 9400 Imager, GE Healthcare). Intensity of the separated protein spots was quantified by Progenesis program (PerkinElmer, MA, US). To compare protein spot intensity between the SLE patient group and the HL group, Cy5-fluorescent intensity of each protein spot was normalized by Cy3-fluorescent intensity of the identical spot. The normalized Cy5-intensity was used for the comparison.
Protein identification
Protein spots of interest, excised from the 2D gels, were digested by trypsin as described previously [15]. Produced peptides were subjected to matrix-assisted laser desorption ionization-time of flight/time of flight mass spectrometry (MALDI-TOF/TOF-MS) (Ultraflex, Bruker Daltonics Bremen, Germany). Determined peptide masses were compiled to allow searches of the National Center for Biotechnology Information (NCBI) protein database using Mascot software program (Matrix Science, London, UK).
Analysis of CS proteins of PBMCs from patients with SLE and HL by 2D-DIGE
To identify SLE-specific CS protein profiles of PBMCs, we comprehensively compared CS proteins of PBMCs between 5 patients with SLE and 5 HLs using 2D-DIGE of isolated CS proteins by cell surface biotinylation. Clinical features of the 5 patients with SLE are described in Table 1.
Table 1 Clinical information of enrolled patients with SLE
Patients |
SLE1 |
SLE2 |
SLE3 |
SLE4 |
SLE5 |
Sex/Age (y.o.) |
F/23 |
F/45 |
F/46 |
F/36 |
F/54 |
Disease duration (y) |
9y |
17y |
1y |
11y |
33y |
Clinical manifestations at the onset and during the course |
Malar rash, Pericarditis, Nephritis, Anti-Sm(+), ANA(+), Plateletpenia, Psychosis |
Non-erosive arthritis,Malar rash, Nephritis,Fever, Skin ulcer,Hyper-CK-emia,Hypocomplementemia |
Non-erosive arthritis,ANA(+), Malar rash,Anti-DNA(+), NephritisHypocomplementemia, |
Fever, anti-DNA(+), ANA(+), Pancytopenia,Nephritis, CNS lesion |
Malar rash, Fever, Pleuritis, Nephritis, ANA(+), Anti-DNA(+) |
Clinical symptoms at the day of sampling |
Malar rash, Discoid rash,Photosensitivity, Psychosis, Nephritis |
Malar rash,Photosensitivity,Non-erosive arthritis |
Malar rash,Non-erosive arthritis |
Malar rash |
Non-erosive arthritis |
Laboratory examinations* |
|
|
|
|
|
WBC (/mL) Lymphocytes (/mL) Hemoglobin (g/dL) Platelets (x103/mL) |
7,00021011.9200 |
7,90089313.7196 |
11,100214212.2204 |
8,10059912.3317 |
6,30076213.3209 |
CH50 (U/mL) |
14.9 |
53.7 |
<10 |
40.9 |
43.9 |
Anti-dsDNA-IgG (IU/mL)(negative ≤10) |
29.7 |
≤10 |
323 |
23.5 |
≤10 |
ANA |
1:320 |
1:2560 |
1:1280 |
1:160 |
1:320 |
SLEDAI* |
18 |
10 |
12 |
8 |
4 |
Treatment* |
PSL (8mg) |
PSL (40mg) |
PSL (20mg) |
PSL (10mg) |
PSL (12mg), Tacrolimus (3mg) |
*Findings and drugs at the day of sampling are shown.
As a result, in total 468 definite protein spots were detected (Figure 1A). We then compared intensity of the 468 protein spots between the SLE patient group and the HL group. We found that 151 (32.3%) out of the 468 protein spots showed significantly different intensity between the two groups (Table 2). Among them, 137 spots (29.3%) showed >±1.5-fold intensity differences between the two groups. Furthermore, 44 spots (9.4%) showed >±2.5-fold intensity differences (Table 2). This study revealed that CS protein profiles of PBMCs in SLE patients were greatly skewed compared to those in HLs.
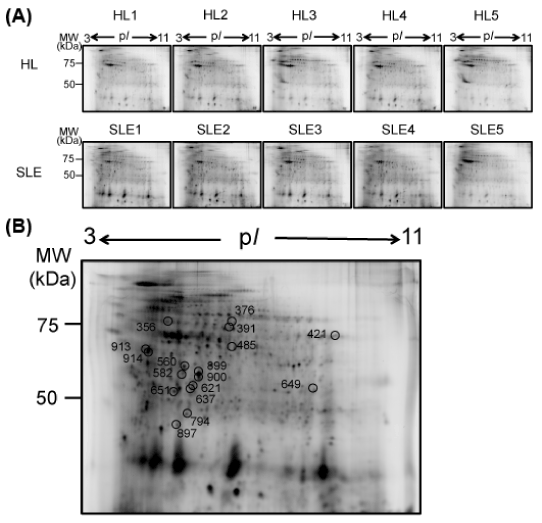
Figure 1. . 2D-DIGE analysis of CS proteins of PBMCs from patients with SLE and HLs. A) CS proteins were extracted from PBMCs obtained from 5 patients with SLE and 5 HLs and then were subjected to 2D-DIGE. Each protein sample labeled with Cy5 and an internal control "standard sample" labeled with Cy3 were mixed and applied onto an IEF gel. The protein samples were separated on 24 cm pH 3-11 non-linear IEF strips and further separated by 12.5% SDS-PAGE. Results from the patients with SLE (Cy5, SLE1-5) and HLs (Cy5, HL1-5) are shown. MW, molecular weight. B) 44 protein spots that showed significantly different intensity between the SLE patient group and the HL group were subjected to the MS/MS analysis to identify proteins. Proteins in 17 out of the 44 protein spots were identified, as summarized in Table 3. Positions of the 17 spots on the 2D gel are shown by circles with spot numbers.
Table 2. Numbers of protein spots that showed significantly different intensity between the SLE patient group and the HL group
Total number of detected protein spots |
Number of spots with
different intensity
( p < 0.05 ) |
Fold difference ( X, SLE / HL ) |
Number of spots |
468 |
151 |
X ≥ 3.5
3.5 > X ≥ 2.5
2.5 > X ≥ 1.5
1.5 > X > -1.5
-1.5 ≥ X > -2.5
-2.5 ≥ X > -3.5
-3.5 ≥ X |
6
23
41
14
52
13
2 |
Identification of proteins with significant intensity differences between the SLE patient group and the HL group: The 44 protein spots that showed > ±2.5-fold intensity differences were subjected to MS/MS analysis to identify protein names (Figure 1B). As a result, proteins in 17 out of the 44 spots were identified as summarized in (Table 3).
Table 3. Protein identification from the protein spots with significantly different intensity between the SLE patient group and the HL group. Peptides recovered from the 17 protein spots after in-gel digestion with trypsin were subjected to MALDI-TOF/TOF-MS analysis. MW; molecular weight, pI; Isoelectric points, LID; laser-induced dissociation
Spot no. |
MW pI
Observed |
Diffrence
(SLE/HL) |
Protein |
Accession no. |
MW pI
Theoretical |
Matched
peptides |
Mascot score |
Coverage (%) |
Sequence confirmed by LID (Mascot ion score) |
649 |
27.3 7.84 |
5.85 |
Myeloblastin (PRTN3_HUMAN) |
gi: 6174926 |
27.8 8.72 |
3 |
73 |
11 |
80LVNNVLGAHNVR91 (9)
228LFPDFFTR235 (16)
236VALYVDWIR244 (26) |
899 |
31.8 6.27 |
3.72 |
Cyclin-L1 (CCNL_HUMAN) |
gi: 74753368 |
59.6 10.7 |
6 |
72 |
13 |
99LPQVAMATGQVLFHR113 (17) |
651 |
27.2 5.07 |
3.24 |
Actin, cytoplasmic 1 (ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN) |
gi: 46397333
gi: 54036678 |
41.7 5.29
41.8 5.31 |
5 |
161 |
21 |
29AVFPSOVGRPR39 (10)
40HQGVMVGMGQK50 (2)
85IWHHTFYNELR95 (73)
96VAPEEHPVLLTEAPLNPK113 (49) |
485 |
40.6 62.7 |
2.90 |
Leukocyte elastase inhibitor (ILEU_HUMAN) |
gi: 266344 |
42.7 5.90 |
3 |
108 |
7 |
57TFHFNTVEEVHSR69 (57) |
|
|
|
|
|
|
|
|
|
276FKLEESYTLNSDLAR290 (28) |
582 |
32 5.26 |
2.82 |
Actin, cytoplasmic 1(ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN) |
gi: 46397333
gi: 4501887 |
41.7 5.29
41.8 5.31 |
6 |
119 |
19 |
239SYELPDGQVITIGNER254 (74)
285CDVDIR290 (5) |
734 |
23.4 5.32 |
2.81 |
Actin, cytoplasmic 1(ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN)
Beta-actin-like protein 2 (ACTBL_HUMAN) |
gi: 46397333
gi: 4501887
gi: 172046825 |
41.7 5.29
41.8 5.31
42.0 5.39 |
3 |
74 |
6 |
239SYELPDGQVITIGNER254 (59)
285CDVDIR290 (2)
240SYELPDGQVITIGNER250(59)
286CDVDIR291(2) |
913 |
40.2 4.56 |
2.81 |
Actin, cytoplasmic 1(ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN) |
gi: 46397333
gi: 4501887 |
41.7 5.29
41.8 5.31 |
4 |
265 |
14 |
29AVFPSIVGRPR39 (37)
85IWHHTFYNELR95 (74)
239SYELPDGQVITIGNER254 (127) |
560 |
34 5.27 |
2.76 |
Actin, cytoplasmic 1(ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN) |
gi: 46397333
gi: 4501887 |
4.17 5.29
41.8 5.31 |
4 |
96 |
14 |
239SYELPDGQVITIGNER254 (83)
285CDVDIR290 (6) |
914 |
40 4.62 |
2.70 |
Actin, cytoplasmic 1 (ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN)
Actin, gamma-enteric smooth muscle (ACTH_HUMAN)
Actin, aortic smooth muscle (ACTA_HUMAN)
Actin, alpha cardiac muscle 1 (ACTC_HUMAN)
Actin, alpha skeletal muscle (ACTS_HUMAN) |
gi: 46397333
gi: 4501887
gi: 54036679
gi: 51316972
gi: 54036697
gi: 61218043 |
41.7 5.29
41.8 5.31
41.9 5.31
42.0 5.23
42.0 5.23
42.0 5.23 |
2 |
110 |
7 |
29AVFPSIVGRPR39 (9)
239SYELPDGQVITIGNER254 (87)
30AVFPSIVGRPR40 (9)
240SYELPDGQVITIGNER255 (87)
31AVFPSIVGRPR41 (9)
241SYELPDGQVITIGNER256 (87) |
897 |
22.1 5.14 |
2.66 |
DNA-directed RNA polymerase II Ⅱ subunitRPB11-b2 (RPB1C_HUMAN) |
gi: 74725032 |
13.1 5.88 |
4 |
56 |
27 |
104FRTCLLPLR112 (9) |
900 |
30 5.57 |
2.66 |
POTE ankyrin domain family member F |
gi: 153791352 |
121 5.83 |
3 |
67 |
4 |
939SYELPDGQVITIGNER954 (59) |
637 |
27.9 5.43 |
2.55 |
Actin, cytoplasmic 1(ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN)
Beta-actin-like protein 2 (ACTBL_HUMAN) |
gi: 46397333
gi: 4501887
gi: 172046825 |
41.7 5.29
41.8 5.31
42.0 5.39 |
3 |
101 |
6 |
239SYELPDGQVITIGNER254 (88) |
621 |
28.7 5.45 |
2.53 |
Putativebeta-actin-likeprotein3(ACTBM_HUMAN)
Actin, cytoplasmic 1 (ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN)
Bate-actin-like protein 2 (ACTBL_HUMAN)
Actin, gamma-enteric smooth muscle (ACTH_HUMAN)
Actin, aortic smooth muscle (ACTA_HUMAN)
Actin, alpha cardiac muscle 1 (ACTC_HUMAN)
Actin, alpha skeletal muscle (ACTS_HUMAN) |
gi: 74739412
gi; 46397333
gi: 4501887
gi: 172046825
gi: 54036679
gi: 51316972
gi: 54036697
gi: 61218043 |
42.0 5.91
41.7 5.29
41.8 5.31
42.0 5.39
41.9 5.31
42.0 5.23
42.0 5.23
42.0 5.23 |
2 |
62 |
4 |
239YELPDGQVITIGNER254 (54)
240YELPDGQVITIGNER255 (54)
241YELPDGQVITIGNER256 (54) |
421 |
47.7 8.28 |
-2.75 |
Meiosis 1 arrest protein (M1AR_HUMAN) |
gi: 74730556 |
59.3 6.40 |
7 |
72 |
17 |
379IPASTFYVIMPSHSLTLLVK398 (22) |
376 |
55.8 6.29 |
-3.09 |
Heterogeneous nuclear ribonucleoprotein H (HNRH1_HUMAN) |
gi: 1710632 |
49.2 5.89 |
7 |
77 |
24 |
300ATENDIYNFFSPLNPVR316 (26) |
391 |
52.9 6.19 |
-3.21 |
Pyruvate kinase PKM (KPYM_HUMAN) |
gi: 20178296 |
57.9 7.96 |
10 |
92 |
21 |
384EAEAAIYHLQLFEELR399 (13) |
356 |
59 4.92 |
-3.40 |
Tubulin alpha-4Achain (TBA4A_HUMAN) |
gi: 55977476 |
49.9 4.95 |
11 |
104 |
30 |
65AVFVDLEPTVIDEIR79 (27) |
Thirteen out of the 17 identified spots showed higher intensity in the SLE patient group than in the HL group (no. 649 of myeloblastin, no. 899 of cyclin-L1, no. 485 of leukocyte elastase inhibitor, no. 897 of DNA-directed RNA polymerase II subunit RPB11-b2, no. 900 of POTE ankyrin domain family member F, and nos. 651, 582, 734, 913, 560, 914, 637, and 621 of actins) (Figure 2, upper and middle). The remaining 4 spots showed lower intensity in the SLE group than in the HL patient group (no. 421 of meiosis 1 arrest protein, no. 376 of heterogeneous nuclear ribonucleoprotein H, no. 391 of pyruvate kinase PKM and no. 356 of tubulin alpha-4A chain) (Figure 2, lower).

Figure 2. Representative protein spots that showed significantly different intensity between the SLE patient group and the HL group.
Representative results from nos. 899, 649, and 391 are shown. 3D image of the protein spot (nos. 649, 899, and 391) was constructed by Progenesis program (Left panel). Normalized intensity of the spot was compared between the SLE patient group and the HL group (Right panel). An average of the normalized protein spot intensity in the HL group was defined as 1.0. A bar shows an average of the normalized protein spot intensity in each group.
Our study revealed that CS protein profiles of PBMCs in patients with SLE were greatly different from those in HLs. Of the differently expressed CS proteins, 10 proteins from 17 proteins spots were identified.
CS proteins have been generally analyzed by FACS. In FACS, targeted CS proteins of live cells are labelled with fluorescence-conjugated target-specific antibodies, after which intensity of bound fluorescence are measured. It is well recognized that FACS is a very effective tool to analyze CS proteins. In fact, FACS has been often used in the analysis of the pathophysiology of SLE [10-13]. However, FACS can analyze only limited kinds (usually 1-4 kinds, at most 8 kinds) of CS proteins. FACS cannot comprehensively analyze CS proteins. In comparison, biotin labeling of CS proteins combined with 2D-DIGE as used here can comprehensively detect and quantitate CS proteins. This is a great advantage of our method. Another point different from FACS is that proteins that bind to biotinylated CS proteins are also isolated together with CS proteins themselves, even if they themselves do not exist on the outside of cell membranes. In this regard, the method used here would be also adequate to analyze translocation of proteins from cytosol or nucleus to the plasma membrane and vice versa. Our strategy and FACS should be differently used depending on aims of studies.
Our study revealed that approximately one-thirds (32.3%) of the detected protein spots showed significantly different expression between the SLE patient group and the HL group. This indicates that CS protein profiles of PBMCs of SLE patients skews very widely. Out of the 44 protein spots with highly different intensity (> ±2.5 folds), proteins of 17 spots were identified, which were composed of 10 kinds of proteins.
First, spot no. 649, which showed 5.85-fold higher intensity in the SLE patient group than in the HL group, was identified as myeloblastin. It is also known as proteinase 3 (PR3). PR3 is a serine protease that is mainly located in granules of neutrophils and eosinophils [16]. PR3 is also known as a target of anti-neutrophil cytoplasmic antibodies (ANCA) [17]. It was reported that mRNA for PR3 in monocytes was increased in patients with SLE compared to HL [18]. Furthermore, it was reported that amounts of cell surface PR3 were increased by TNF-stimulation in neutrophils of patients with SLE and monocytes of patients with ANCA-associated vasculitis [19,20]. In addition, serum TNF- levels were reported to be increased in patients with SLE [21]. Taking our data together with these reports, both PR3 expression and its translocation to the plasma membrane fraction would be increased in PBMCs of patients with SLE. Roles of the increase in the pathogenesis of SLE should be investigated in the future.
Second, spot no. 899, which showed 2,53~3,24-fold higher intensity in the SLE patient group than in the HL group, was identified as cyclin L1. Cyclin L1 is usually located in cytoplasm. It was reported that cyclin L1 was involved in the splicing of mRNA [22,23]. In T cells of patients with SLE, abnormal splicing of TCR zeta chain was reported [24,25]. Our data and their reports indicate possible involvement of translocation of cyclin L1 in the abnormal splicing in T cells of patients with SLE. This point should be also investigated in detail in the future.
2021 Copyright OAT. All rights reserv
Third, spot nos. 560, 582, 621, 637, 651, 734, 913, and 914, intensity of which was 2.53~3.24 fold higher in the SLE patient group than in the HL group, were assigned to members of the actin family. The existence of multiple spots would indicate different isoforms of actin and/or different post-translational modification of actin. Since actin is an important component of cytoskeleton, PBMCs in patients with SLE would alter the structure of cytoskeleton, reflecting functional abnormalities of lymphocytes. Further studies are needed to clarify the relation between the actin alteration and functional abnormalities of lymphocytes.
In conclusion, our study comprehensively investigated CS protein profiles of PBMCs from patients with SLE for the first time, to our knowledge. We found that CS protein profiles from PBMCs from patients with SLE are greatly different from those of HLs. Our study would provide a new strategy to investigate the pathophysiology of SLE.
We thank Ms. Yokoyama MK for her technical assistance.
The authors has no conflicts of interest.
- Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, et al. (1982) The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 25: 1271-1277. [Crossref]
- Yu C, Gershwin ME, Chang C (2014) Diagnostic criteria for systemic lupus erythematosus: a critical review. J Autoimmun 48-49: 10-3. [Crossref]
- Petri M, Orbai AM, Alarcon GS, Gordon C, Merrill JT, et al. (2012) Derivation and validation of the systemic lupus international collaborating clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum 64: 2677–2686. [Crossref]
- Miranda-Carús ME, Askanase AD, Clancy RM, Di Donato F, Chou TM, et al. (2000) Anti-SSA/Ro and anti-SSB/La autoantibodies bind the surface of apoptotic fetal cardiocytes and promote secretion of TNF-alpha by macrophages. J Immunol. 165: 5345-5351.
- Cozzani E, Agnoletti AF, Pappalardo F, Schiavetti I, Torino A, et al. (2016) The high incidence of anti-Ro/SSA and anti-p200 antibodies in female patients with connective tissue diseases confirms the importance of screening for congenital heart block-associated autoantibodies during pregnancy. Arch Dermatol Res. 308: 139-143.
- Mak A, Kow NY (2014) The pathology of T cells in systemic lupus erythematosus. J Immunol Res 2014: 419029. [Crossref]
- Shlomchik MJ, Craft JE, Mamula MJ (2001) From T to B and back again: positive feedback in systemic autoimmune disease. Nat Rev Immunol 1: 147-153. [Crossref]
- Anolik JH (2013) B cell biology: implications for treatment of systemic lupus erythematosus. Lupus 22: 342-349. [Crossref]
- Kow NY, Mak A. (2013) Costimulatory pathways: physiology and potential therapeutic manipulation in systemic lupus erythematosus. Clin Dev Immunol 16: 245928.
- Hase K, Tani K, Shimizu T, Ohmoto Y, Matsushima K, et al. (2001) Increased CCR4 expression in active systemic lupus erythematosus. J Leukoc Biol 70: 749-755. [Crossref]
- Bohgaki T, Amasaki Y, Nishimura N, Bohgaki M, Yamashita Y, et al. (2005) Up regulated expression of tumour necrosis factor α converting enzyme in peripheral monocytes of patients with early systemic sclerosis. Ann Rheum Dis 64: 1165-1173.
- Nakano S, Morimoto S, Suzuki J, Nozawa K, Amano H, et al. (2008) Role of pathogenic auto-antibody production by Toll-like receptor 9 of B cells in active systemic lupus erythematosus. Rheumatology (Oxford) 47: 145-149.
- Herrada AA, Llanos C, Mackern-Oberti JP, Carreño LJ, Henriquez C, et al. (2012) Haem oxygenase 1 expression is altered in monocytes from patients with systemic lupus erythematosus. Immunology 136: 414-424.
- Onodera H, Arito M, Sato T, Ito H, Hashimoto T, et al. (2013) Novel effects of edaravone on human brain microvascular endothelial cells revealed by a proteomic approach. Brain Res 1534: 87-94. [Crossref]
- Fujisawa H, Ohtani-Kaneko R, Naiki M, Okada T, Masuko K, et al. (2008) Involvement of post-translational modification of neuronal plasticity-related proteins in hyperalgesia revealed by a proteomic analysis. Proteomics 8: 1706-1719.
- Kettritz R (2016) Neutral serine proteases of neutrophils. Immunol Rev 273: 232-248. [Crossref]
- van der Geld YM, Limburg PC, Kallenberg CG (2001) Proteinase 3, Wegener's autoantigen: from gene to antigen. J Leukoc Biol 69: 177-190. [Crossref]
- Ohlsson S, Hellmark T, Pieters K, Sturfelt G, Wieslander J, et al. (2005) Increased monocyte transcription of the proteinase 3 gene in small vessel vasculitis. Clin Exp Immunol 141: 174-182. [Crossref]
- van Rossum AP, Huitema MG, Stegeman CA, Bijl M, de Leeuw K, et al. (2007) Standardised assessment of membrane proteinase 3 expression. Analysis in ANCA-associated vasculitis and controls. Ann Rheum Dis 66: 1350-1355.
- Hattar K, Bickenbach A, Csernok E, Rosseau S, Grandel U, et al. (2002) Wegener's granulomatosis: antiproteinase 3 antibodies induce monocyte cytokine and prostanoid release-role of autocrine cell activation. J Leukoc Biol 71: 996-1004. [Crossref]
- Weckerle CE, Mangale D, Franek BS, Kelly JA, Kumabe M, et al. (2012) Large-scale analysis of tumor necrosis factor α levels in systemic lupus erythematosus. Arthritis Rheum 64: 2947-2952.
- Chen HH, Wang YC, Fann MJ (2006) Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Mol Cell Biol 26: 2736-2745.
- Loyer P, Trembley JH, Grenet JA, Busson A, Corlu A, et al. (2008) Characterization of cyclin L1 and L2 interactions with CDK11 and splicing factors: influence of cyclin L isoforms on splice site selection. J Biol Chem. 283: 7721-7732.
- Tsuzaka K, Onoda N, Yoshimoto K, Setoyama Y, Suzuki K, et al. (2002) T-cell receptorζ mRNA with an alternatively spliced 3' untranslated region is generated predominantly in the peripheral blood T cells of systemic lupus erythematosus patients. Mod Rheumatol 12: 167-173.
- Tsuzaka K, Nozaki K, Kumazawa C, Shiraishi K, Setoyama Y, et al. (2006) TCR 3 mRNA splice variant forms observed in the peripheral blood T cells from systemic lupus erythematosus patients. Springer Semin Immun 28: 185-193.
Editorial Information
Editor-in-Chief
Ivan Gout
University College London
Article Type
Research Article
Publication history
Received date: November 20, 2016
Accepted date: December 06, 2016
Published date: December 12, 2016
Copyright
©2016 Arito M. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation
Arito M (2016) Comprehensive analysis of surface proteins of peripheral mononuclear blood cells in patients with systemic lupus erythematosus. Integr Mol Med 3: DOI: 10.15761/IMM.1000260.
Corresponding author
Mitsumi Arito
Clinical Proteomics and Molecular Medicine, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae, Kawasaki, Kanagawa, 216-8511, Japan

Figure 1. . 2D-DIGE analysis of CS proteins of PBMCs from patients with SLE and HLs. A) CS proteins were extracted from PBMCs obtained from 5 patients with SLE and 5 HLs and then were subjected to 2D-DIGE. Each protein sample labeled with Cy5 and an internal “control standard” sample labeled with Cy3 were mixed and applied onto an IEF gel. The protein samples were separated on 24 cm pH 3-11 non-linear IEF strips and further separated by 12.5% SDS-PAGE. Results from the patients with SLE (Cy5, SLE1-5) and HLs (Cy5, HL1-5) are shown. MW, molecular weight. B) 44 protein spots that showed significantly different intensity between the SLE patient group and the HL groups were subjected to the MS/MS analysis to identify proteins. Proteins in 17 out of the 44 protein spots were identified, as summarized in Table 3. Positions of the 17 spots on the 2D gel are shown by circles with spot numbers.

Figure 2. Representative protein spots that showed significantly different intensity between the SLE patient group and the HL group.
Representative results from nos. 899, 649, and 391 are shown. 3D image of the protein spot (nos. 649, 899 and 391) was constructed by Progenesis program (Left panel). Normalized intensity of the spot was compared between the SLE patient group and the HL group (Right panel). An average of the normalized protein spot intensity in the HL group was defined as 1.0. A bar shows an average of the normalized protein spot intensity in each group.
Table 1 Clinical information of enrolled patients with SLE
Patients |
SLE1 |
SLE2 |
SLE3 |
SLE4 |
SLE5 |
Sex/Age (y.o.) |
F/23 |
F/45 |
F/46 |
F/36 |
F/54 |
Disease duration (y) |
9y |
17y |
1y |
11y |
33y |
Clinical manifestations at the onset and during the course |
Malar rash, Pericarditis, Nephritis, Anti-Sm(+), ANA(+), Plateletpenia, Psychosis |
Non-erosive arthritis,Malar rash, Nephritis,Fever, Skin ulcer,Hyper-CK-emia,Hypocomplementemia |
Non-erosive arthritis,ANA(+), Malar rash,Anti-DNA(+), NephritisHypocomplementemia, |
Fever, anti-DNA(+), ANA(+), Pancytopenia,Nephritis, CNS lesion |
Malar rash, Fever, Pleuritis, Nephritis, ANA(+), Anti-DNA(+) |
Clinical symptoms at the day of sampling |
Malar rash, Discoid rash,Photosensitivity, Psychosis, Nephritis |
Malar rash,Photosensitivity,Non-erosive arthritis |
Malar rash,Non-erosive arthritis |
Malar rash |
Non-erosive arthritis |
Laboratory examinations* |
|
|
|
|
|
WBC (/mL) Lymphocytes (/mL) Hemoglobin (g/dL) Platelets (x103/mL) |
7,00021011.9200 |
7,90089313.7196 |
11,100214212.2204 |
8,10059912.3317 |
6,30076213.3209 |
CH50 (U/mL) |
14.9 |
53.7 |
<10 |
40.9 |
43.9 |
Anti-dsDNA-IgG (IU/mL)(negative ≤10) |
29.7 |
≤10 |
323 |
23.5 |
≤10 |
ANA |
1:320 |
1:2560 |
1:1280 |
1:160 |
1:320 |
SLEDAI* |
18 |
10 |
12 |
8 |
4 |
Treatment* |
PSL (8mg) |
PSL (40mg) |
PSL (20mg) |
PSL (10mg) |
PSL (12mg), Tacrolimus (3mg) |
*Findings and drugs at the day of sampling are shown.
Table 2. Numbers of protein spots that showed significantly different intensity between the SLE patient group and the HL group
Total number of detected protein spots |
Number of spots with
different intensity
( p < 0.05 ) |
Fold difference ( X, SLE / HL ) |
Number of spots |
468 |
151 |
X ≥ 3.5
3.5 > X ≥ 2.5
2.5 > X ≥ 1.5
1.5 > X > -1.5
-1.5 ≥ X > -2.5
-2.5 ≥ X > -3.5
-3.5 ≥ X |
6
23
41
14
52
13
2 |
Table 3. Protein identification from the protein spots with significantly different intensity between the SLE patient group and the HL group. Peptides recovered from the 17 protein spots after in-gel digestion with trypsin were subjected to MALDI-TOF/TOF-MS analysis. MW; molecular weight, pI; Isoelectric points, LID; laser-induced dissociation
Spot no. |
MW pI
Observed |
Diffrence
(SLE/HL) |
Protein |
Accession no. |
MW pI
Theoretical |
Matched
peptides |
Mascot score |
Coverage (%) |
Sequence confirmed by LID (Mascot ion score) |
649 |
27.3 7.84 |
5.85 |
Myeloblastin (PRTN3_HUMAN) |
gi: 6174926 |
27.8 8.72 |
3 |
73 |
11 |
80LVNNVLGAHNVR91 (9)
228LFPDFFTR235 (16)
236VALYVDWIR244 (26) |
899 |
31.8 6.27 |
3.72 |
Cyclin-L1 (CCNL_HUMAN) |
gi: 74753368 |
59.6 10.7 |
6 |
72 |
13 |
99LPQVAMATGQVLFHR113 (17) |
651 |
27.2 5.07 |
3.24 |
Actin, cytoplasmic 1 (ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN) |
gi: 46397333
gi: 54036678 |
41.7 5.29
41.8 5.31 |
5 |
161 |
21 |
29AVFPSOVGRPR39 (10)
40HQGVMVGMGQK50 (2)
85IWHHTFYNELR95 (73)
96VAPEEHPVLLTEAPLNPK113 (49) |
485 |
40.6 62.7 |
2.90 |
Leukocyte elastase inhibitor (ILEU_HUMAN) |
gi: 266344 |
42.7 5.90 |
3 |
108 |
7 |
57TFHFNTVEEVHSR69 (57) |
|
|
|
|
|
|
|
|
|
276FKLEESYTLNSDLAR290 (28) |
582 |
32 5.26 |
2.82 |
Actin, cytoplasmic 1(ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN) |
gi: 46397333
gi: 4501887 |
41.7 5.29
41.8 5.31 |
6 |
119 |
19 |
239SYELPDGQVITIGNER254 (74)
285CDVDIR290 (5) |
734 |
23.4 5.32 |
2.81 |
Actin, cytoplasmic 1(ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN)
Beta-actin-like protein 2 (ACTBL_HUMAN) |
gi: 46397333
gi: 4501887
gi: 172046825 |
41.7 5.29
41.8 5.31
42.0 5.39 |
3 |
74 |
6 |
239SYELPDGQVITIGNER254 (59)
285CDVDIR290 (2)
240SYELPDGQVITIGNER250(59)
286CDVDIR291(2) |
913 |
40.2 4.56 |
2.81 |
Actin, cytoplasmic 1(ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN) |
gi: 46397333
gi: 4501887 |
41.7 5.29
41.8 5.31 |
4 |
265 |
14 |
29AVFPSIVGRPR39 (37)
85IWHHTFYNELR95 (74)
239SYELPDGQVITIGNER254 (127) |
560 |
34 5.27 |
2.76 |
Actin, cytoplasmic 1(ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN) |
gi: 46397333
gi: 4501887 |
4.17 5.29
41.8 5.31 |
4 |
96 |
14 |
239SYELPDGQVITIGNER254 (83)
285CDVDIR290 (6) |
914 |
40 4.62 |
2.70 |
Actin, cytoplasmic 1 (ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN)
Actin, gamma-enteric smooth muscle (ACTH_HUMAN)
Actin, aortic smooth muscle (ACTA_HUMAN)
Actin, alpha cardiac muscle 1 (ACTC_HUMAN)
Actin, alpha skeletal muscle (ACTS_HUMAN) |
gi: 46397333
gi: 4501887
gi: 54036679
gi: 51316972
gi: 54036697
gi: 61218043 |
41.7 5.29
41.8 5.31
41.9 5.31
42.0 5.23
42.0 5.23
42.0 5.23 |
2 |
110 |
7 |
29AVFPSIVGRPR39 (9)
239SYELPDGQVITIGNER254 (87)
30AVFPSIVGRPR40 (9)
240SYELPDGQVITIGNER255 (87)
31AVFPSIVGRPR41 (9)
241SYELPDGQVITIGNER256 (87) |
897 |
22.1 5.14 |
2.66 |
DNA-directed RNA polymerase II Ⅱ subunitRPB11-b2 (RPB1C_HUMAN) |
gi: 74725032 |
13.1 5.88 |
4 |
56 |
27 |
104FRTCLLPLR112 (9) |
900 |
30 5.57 |
2.66 |
POTE ankyrin domain family member F |
gi: 153791352 |
121 5.83 |
3 |
67 |
4 |
939SYELPDGQVITIGNER954 (59) |
637 |
27.9 5.43 |
2.55 |
Actin, cytoplasmic 1(ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN)
Beta-actin-like protein 2 (ACTBL_HUMAN) |
gi: 46397333
gi: 4501887
gi: 172046825 |
41.7 5.29
41.8 5.31
42.0 5.39 |
3 |
101 |
6 |
239SYELPDGQVITIGNER254 (88) |
621 |
28.7 5.45 |
2.53 |
Putativebeta-actin-likeprotein3(ACTBM_HUMAN)
Actin, cytoplasmic 1 (ACTB_HUMAN)
Actin, cytoplasmic 2 (ACTG_HUMAN)
Bate-actin-like protein 2 (ACTBL_HUMAN)
Actin, gamma-enteric smooth muscle (ACTH_HUMAN)
Actin, aortic smooth muscle (ACTA_HUMAN)
Actin, alpha cardiac muscle 1 (ACTC_HUMAN)
Actin, alpha skeletal muscle (ACTS_HUMAN) |
gi: 74739412
gi; 46397333
gi: 4501887
gi: 172046825
gi: 54036679
gi: 51316972
gi: 54036697
gi: 61218043 |
42.0 5.91
41.7 5.29
41.8 5.31
42.0 5.39
41.9 5.31
42.0 5.23
42.0 5.23
42.0 5.23 |
2 |
62 |
4 |
239YELPDGQVITIGNER254 (54)
240YELPDGQVITIGNER255 (54)
241YELPDGQVITIGNER256 (54) |
421 |
47.7 8.28 |
-2.75 |
Meiosis 1 arrest protein (M1AR_HUMAN) |
gi: 74730556 |
59.3 6.40 |
7 |
72 |
17 |
379IPASTFYVIMPSHSLTLLVK398 (22) |
376 |
55.8 6.29 |
-3.09 |
Heterogeneous nuclear ribonucleoprotein H (HNRH1_HUMAN) |
gi: 1710632 |
49.2 5.89 |
7 |
77 |
24 |
300ATENDIYNFFSPLNPVR316 (26) |
391 |
52.9 6.19 |
-3.21 |
Pyruvate kinase PKM (KPYM_HUMAN) |
gi: 20178296 |
57.9 7.96 |
10 |
92 |
21 |
384EAEAAIYHLQLFEELR399 (13) |
356 |
59 4.92 |
-3.40 |
Tubulin alpha-4Achain (TBA4A_HUMAN) |
gi: 55977476 |
49.9 4.95 |
11 |
104 |
30 |
65AVFVDLEPTVIDEIR79 (27) |

