Abstract
Smoking and infection with Human Papilloma virus (HPV) are important risk factors in head and neck squamous cell carcinoma (HNSCC). In spite of the fact that HPV-associated HNSCC patients tend to respond better to current treatment strategies than non-HPV associated HNSCC, some patients with HPV-associated HNSCC develop disease recurrences and develop distant metastases, which can be resistant to current treatment strategies. Cancer stem cells (CSCs) are believed to underlie the development of this resistant phenotype. In this study we examined the expression of CSC markers genes, as well as that of the STAT3 and Notch pathway genes in HPV-associated HNSCC patients, which we previously found to be overexpressed in glioma CSCs. Xenografts were established in immunocompromised mice from two patients (UTSCC-1 and UTSCC-2) with HPV-associated HNSCC that differed in their history of tobacco use. RNA extracted from both tumors showed high expression of SOX2, Nestin, CD133 and CD44, but the UTSCC-2 tumor from a patient with a history of heavy tobacco use showed significantly overexpressed SOX2, CD44 and CD133 compared to UTSCC-1. Moreover, UTSCC-2 significantly overexpressed many Notch pathway genes. Immunochemistry of STAT3 and pSTAT3 also showed strong expression of STAT3 and pSTAT3 in UTSCC-2 tumor. Serial transplantation of UTSCC-1 enhanced tumor formation, resulted in lung metastasis, and overexpression of STAT3 and NOTCH pathway genes. In conclusion, genes implicated in CSCs, STAT3 and NOTCH pathways are overexpressed in HPV-associated HNSCC, suggesting that these pathways may serve as novel targets for therapeutic intervention for this subset of patients.
Key words
cancer stem cells, head and neck squamous cell carcinoma, human papilloma virus, Notch, STAT3
Abbreviations
CSCs: Cancer stem cells; HNSCC: head and neck squamous cell carcinoma; HPV: human Papilloma virus; NSG: Prkdcscid Il2rgtm1Wjl/; TCGA: The Cancer Genome Atlas; pSTAT3: tyrosine phosphorylated STAT3
Introduction
Head and Neck Squamous Cell Cancer (HNSCC) is the sixth most common cancer, but treatment advances over the past three decades have not affected the overall 5-year survival [1]. A history of smoking is an important risk factor associated with the development of HNSCC [2]. In addition, there has been a sharp rise in the incidence of HPV-HNSCC, which has surpassed the incidence of HPV-induced cervical cancer [3,4]. However, although patients with HPV-HNSCC fare better than those with non-HPV associated HNSCC, a significant subset of HPV-HNSCC patients eventually develops recurrent disease as well as metastases. Tobacco exposure has been proposed to explain in part why some patients with HPV- HNSCC have a worse prognosis [5].
Cancer stem cells (CSCs) have been hypothesized to play an important role in treatment resistance and tumor recurrence in many solid tumors, including HNSCC [6]. CSCs represent a small subpopulation of tumor cells that express a number of stem cell markers [7,8]. CSCs have the unique ability to self-renew and form differentiated progeny, which results in tumors composed of a heterogeneous cell population [6,9]. Tumor cells that undergo epithelial to mesenchymal transition represent a key population of cells that lead to formation of metastases [10]. Interestingly, the CSC enriched population in glioblastoma represents CSCs undergoing the mesenchymal transition [11]. CSCs show elevated expression of CD44 and CD133, which are classic stem cell markers. Interestingly, these CSCs upregulate several Notch pathway genes due to constitutive activation of the STAT3 transcription factor [12]. STAT3 is activated through tyrosine phosphorylation (pSTAT3) by a wide variety of cytokines and growth factors suggesting that it integrates diverse signals into common transcriptional responses [13-15]. High persistent activation of STAT3 is found in HNSCC [16], and apparently actively participates in tumor formation and progression [17]. Notch signaling appears to play a key role in CSCs in a variety of cancers, and controls cell fate determination, survival, proliferation and the maintenance of stem cells [18].
Patients with HPV-HNSCC have shown an improved response rate to current treatment modalities when compared to HNSCC patients without any detectable HPV infection. However patients with HPV-HNSCC with a history of significant tobacco use have been found to fare worse than non-tobacco using patients [3]. Despite the improved prognosis of patients with HPV-HNSCC, recent clinical studies have suggested that metastatic and recurrent HPV-HNSCC is more resistant to treatment, and therefore requires multiple modalities for achieving disease control [19] Therefore, identifying the factors that contribute to treatment resistance is of utmost importance in this patient population. In the present study we explored the hypothesis that HPV-HNSCC patients have a more aggressive phenotype due to an increase in CSC populations of their tumor, and that may be affected by their history of tobacco use. We established two xenograft lines in immunocompromised mice from HPV-HNSCC patients that differed in their tobacco use, and examined the expression of genes related to CSCs, which included cell surface markers, and the STAT3 and Notch pathways. These findings could provide information for the development of novel molecular targeted agents for a particular subset of HNSCC patients.
Materials and methods
Patient material
All studies were approved by the University of Tennessee Health Sciences Institutional Review Board. The two study patients had previously untreated oropharyngeal squamous cell carcinoma. HPV etiology was inferred from positive p16 immunostaining. After obtaining informed consent, fresh tumor tissue was obtained during surgical resection of the primary oropharyngeal tumor. Table 1 shows patient characteristics including demographics, staging, pathology, treatment and follow up.
| |
UTSCC-1
|
UTSCC-2
|
|
Patient gender
|
Male
|
Male
|
|
Patient age
|
74 years
|
63 years
|
|
Tumor staging
|
T1N0M0
|
T1N0M0
|
|
Pathology
|
Perineural invasion: no
Lymphovascular invasion: no
Surgical margins: R0 resection
|
Perineural invasion: no
Lymphovascular invasion: no
Surgical margins: R1 resection
|
|
Postoperative treatment
|
None
|
None
|
|
Length of follow up
|
8 months
|
6 months
|
|
Status at follow up
|
No evidence of disease
|
No evidence of disease
|
|
Tobacco Use
|
Less than 10 pack years, cessation 40 years prior (low risk)
|
25 pack years, cessation 1 year prior (high risk)
|
Table 1. Patient demographics.
Growth of bulk HNSCC cell lines and Cancer Stem Cells (CSCs)
Patient tumor samples were washed extensively in DMEM containing antibiotics [penicillin, streptomycin and amphotericin B (Gibco)], and the tumor tissue was minced with surgical blades. For growth in cell culture, the minced tumor tissue was passed through a 70 mm sieve and cultured at 37oC with 5% CO2 in DMEM with 10% fetal bovine serum (Atlanta Biologics) supplemented with penicillin and streptomycin. For CSC enrichment, cells were grown in Neuralbasal media supplemented with B27, epidermal growth factor, fibroblast growth factor and antibiotics on plates coated with poly-D-lysine and laminin.
Gene expression analysis
Total RNA was isolated from homogenized tumor tissue using TRIzol Reagent (Ambion). Real time RT-PCR was performed in triplicate using an iScript One-Step RT-PCR kit with SYBR Green (Bio-Rad). Reaction parameters were as follows: cDNA synthesis at 50°C for 20 min, transcriptase inactivation at 95°C for 5 min, PCR cycling at 95°C for 10 sec, and 60°C for 30 sec for 40 cycles [12]. Gene expression was normalized relative to beta-actin expression. The data presented represents the mean ± standard deviation (SD). Student’s t-tests and ANOVA were performed using GraphPad Prism 4 (GraphPad Software, Inc). p values < 0.05 (*) were considered statistically significant.
The following primers were used for RT-PCR: hACTB: 5’-CATGTACGTTGCTATCCAGGC -3’ (forward) and 5’-CTCCTTAATGTCACGCACGAT -3’(reverse); hSOX2: 5’- GCCGAGTGGAAACTTTTGTCG -3’ (forward) and 5’- GCAGCGTGTACTTATCCTTCTT -3’ (reverse); hNestin: 5’- GGCGCACCTCAAGATGTCC -3’ (forward) and 5’- CTTGGGGTCCTGAAAGCTG (reverse); hCD133: 5’-AGTCGGAAACTGGCAGATAGC-3’ (forward) and 5’- GGTAGTGTTGTACTGGGCCAAT (reverse); hCD44: 5’-GGCACCACTGCTTATGAAGGA-3’ (forward) and 5’ -ACTAGGAGTTGCCTGGATTGT -3’ (reverse); hNOTCH1: 5’-GAGGCGTGGCAGACTATGC-3’ (forward) and 5’-CTTGTACTCCGTCAGCGTGA-3’ (reverse); hNOTCH2: 5’-ATCCCACAAAGCCTAGCACC-3’ (forward) and 5’-CCTTGTCCCTGAGCAACCAT-3’ (reverse); hNOTCH3: 5’-TGTGCAAATGGAGGTCGTT -3’ (forward) and 5’- CCTGAGTGACAGGGGTCCT-3’ (reverse); hNOTCH4: 5’-CAGCCCAGTGGGTATCTCTG -3’ (forward) and 5’- GTTGTGACAGGGTTGGGACT -3’ (reverse); hDTX3: 5’-TCGTTCGTCCTGTCCAGAATG-3’(forward) and 5’-AAGTCTCGCCATCTATGAGGAT-3’(reverse); hNUMBL: 5’ TGGTGGACGACAAAACCAAGG-3’ (forward) and 5’- ACGACAGATATAGGAGAAAGCCT-3’(reverse); hHES: 5’- AGTCCCAAGGAGAAAAACCGA -3’(forward) and 5’- GCTGTGTTTCAGGTAGCTGAC-3’(reverse); hJAG1: 5’-GTCCATGCAGAACGTGAACG-3’(forward) and 5’- GCGGGACTGATACTCCTTGA-3’(reverse); hSTAT3: 5’- CAGCAGCTTGACACACGGTA-3’ (forward) and 5’- GCCCAATCTTGACTCTCAATCC-3’(reverse).
Immunofluorescence and Confocal Microscopy
Tumor tissue was sectioned using a cryostat for indirect immunofluorescence. After washing with PBS, slides were fixed with 4% paraformaldehyde for 15 min at 25°C, followed by an additional three washes with PBS. Slides were then blocked with 5% bovine serum albumin and 1% goat serum in PBS /Triton-X100 at 22oC for 1 hour. Slides were washed 3 times with PBS for 10 minutes, and immunostained with primary antibodies against STAT3 (BD Transduction Laboratories) and pSTAT3 (Abcam) by overnight incubation at 4°C at 1:50 dilution and 1:100 dilution, respectively. The slides were washed 3 times with PBS for 10 min and then secondary antibody, goat anti-rabbit Alexa Fluor 488, was applied and incubated at 25°C for an additional 90 min. DNA was counterstained with Vectashield mounting media with 4′,6-diamidino-2-phenylindole dihydrochloride, DAPI (Vectra Laboratories). Images were captured on a Zeiss LSM700 laser scanning confocal microscope.
Tumor formation in mice
Animal experiments were performed in accordance with a study protocol approved by the Institutional Animal Care and Use Committee of the University of Tennessee Health Science Center. Xenografts were established in five-week old male Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratory) by direct flank injection of 1 × 106 cells from minced tumor tissue after passage through a 70µm sieve. Tumors were measured weekly with a handheld caliper, and harvested when tumor had reached 10 mm. Tumor tissue was fixed in 10% neutral buffered formalin for 24 hours, embedded in paraffin wax, sectioned at 5 µm thickness, and stained with hematoxylin and eosin.
The Cancer Genome Atlas (TCGA) data query
To examine the relationship between STAT3 expression and tumor stage in human cancer specimens from head and neck cancer, we queried the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp) for all HNSCC samples with gene expression data available, as well as the accompanying clinical data. The data set was filtered for samples having STAT3 expression data and clinical data, yielding a final dataset of 521 independent patient samples. Statistical analysis was performed using Graphpad Prism. p values < 0.05 (*) were considered statistically significant.
Results
The growth of HNSCC tumor xenografts
Freshly isolated patient tumor tissue was minced and then directly injected into the flanks of severely immunocompromised NSG mice that are highly sensitive to tumor induction. Although both HNSCC tumor samples formed tumors, tumor growth was rather slow. For example, the UTSCC-1 tumor took 279 days to reach 10 mm, while the UTSCC-2 tumor took 126 days. Histological analysis of tumor tissue showed that both tumor samples were invasive carcinoma, with UTSCC-2 being comprised of less differentiated cells.
CSC marker gene expression in HNSCC tumor tissue
To define whether the tumors expressed evidence of CSCs, we measured the levels of several classical stem cell markers. In brief, total RNA was prepared from the two (UTSCC-1 and UTSCC-2) xenograft tumors and the expression of Sox2, Nestin, CD133 and CD44 was determined by qPCR. As shown in Figure 1, the expression of these CSC marker genes varied markedly between these two HNSCC tumors. The gene expression of stem cell markers SOX2, CD133 and CD44, was significantly increased in UTSCC-2 tumor tissue (p=0.001, p=0.033 and p=0.047, respectively) as compared to their expression in UTSCC-1. Although Nestin gene expression was slightly increased in UTSCC-2 tumor tissue, the difference was not statistically significant (p=0.074). Nonetheless, these results show that several CSC marker genes were expressed at markedly higher levels in UTSCC-2.
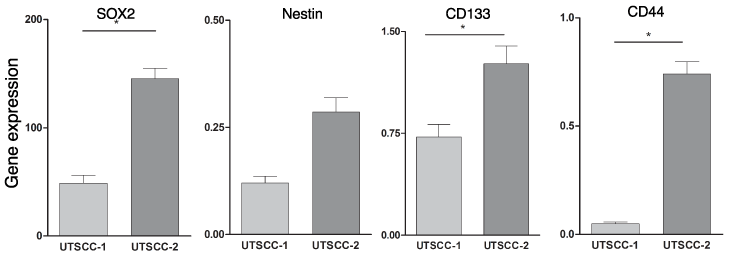
Figure 1. CSC marker gene expression in HNSCC tumor tissue. Total RNA was prepared from UTSCC-1 and UTSCC-2 tumor tissue and gene expression was determined by qPCR, and normalized to actin expression
Expression of genes in the Notch pathway in HNSCC tumor tissue
Since genes in the Notch signaling pathway are enriched in CSCs derived from various tumor types, we determined the expression of various genes in the Notch pathway in the two different HNSCC tumor xenografts. As shown in Figure 2, there was a statistically significant increase in NOTCH1, NOTCH2, NOTCH3 and NOTCH4 expression in UTSCC-2 tumor tissue relative to UTSCC-1 tumor tissue (p=0.027, p=0.018 ,p=0.004 and p=0.0005, respectively). In addition, other Notch pathway genes, such as JAG1, DTX3, and NUMBL are significantly upregulated in UTSCC-2 tumor relative to UTSCC-1 tumor tissue (p=0.008, p=0.0015 and p=0.018 respectively). However, expression of HES5, which plays an important role in Notch pathway, was similar in the two different HNSCC tumors.
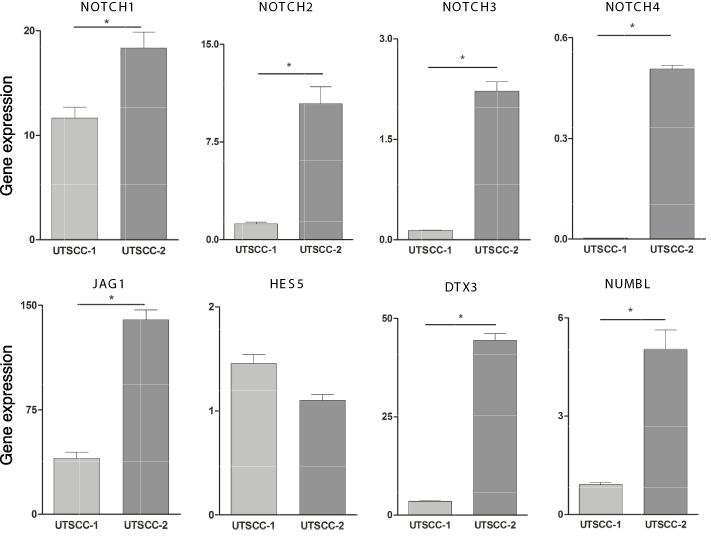
Figure 2. Expression of genes in the Notch pathway in HNSCC tumor tissue. Total RNA was prepared from UTSCC-1 and UTSCC-2 tumor tissue and gene expression was determined by qPCR, and normalized to actin expression
STAT3 gene expression in HNSCC tumor tissue
Since in previous studies we found that STAT3 was constitutively activated as evidenced by its tyrosine phosphorylation in glioma CSCs, and STAT3 gene expression was upregulated [11], we examined STAT3 expression in HNSCC tumor xenografts. As shown in Figure 3A, there was a statistically significant increase (p=0.003) in STAT3 expression in UTSCC-2 tumor tissue relative to UTSCC-1. HNSCC tumor tissue was immunostained for STAT3 and pSTAT3. As shown in Figure 3B, although both STAT3 and pSTAT3 are detectable in both UTSCC-1 and UTSCC-2 tumor tissue, immunostaining for both STAT3 and pSTAT3 was clearly more intense in UTSCC-2 tissue, which is consistent with our qPCR results on STAT3 gene expression. Most importantly, pSTAT3 and STAT3 are selectively colocalized in the nucleus to a greater extent in UTSCC-2 tumor tissue. It is important to note that pSTAT3 and STAT3 colocalization is evident in specific regions in the HNSCC tumor tissue, specifically adjacent to heavily vascularized regions of the tumor.
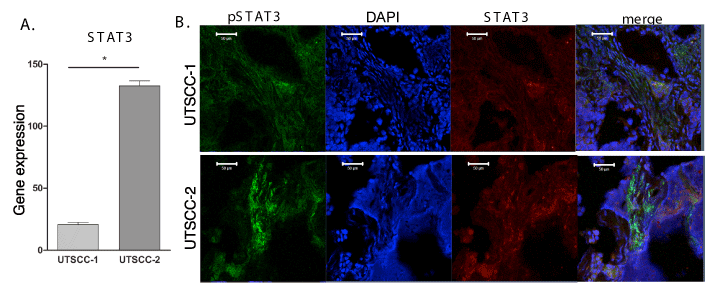
Figure 3. STAT3 gene expression in HNSCC tumor tissue. (A) Total RNA was prepared form UTSCC-1 and UTSCC-2 tumor tissue and STAT3 gene expression was determined by qPCR, and normalized to actin expression. (B) Immunohistochemistry for STAT3 and pSTAT3 in both UTSCC-1 and UTSCC-2 tumor tissue.
2021 Copyright OAT. All rights reserv
We then interrogated the publicly available TCGA database for STAT3 gene expression in HNSCC. The specimens (521 specimens) in the database included all stages of disease varying from lower-stage tumors with only primary site involvement to higher-stage tumors with more advanced status involving lymph nodes. As shown in Figure 4, relatively low STAT3 expression was found in the localized and the least aggressive form of HNSCC (Stage 0 and I), while relatively high STAT3 expression was found in more aggressive HNSCC (Stages II, III and IV). These results provide additional evidence that STAT3 may play an important role in HNSCC tumorigenesis and aggressiveness.
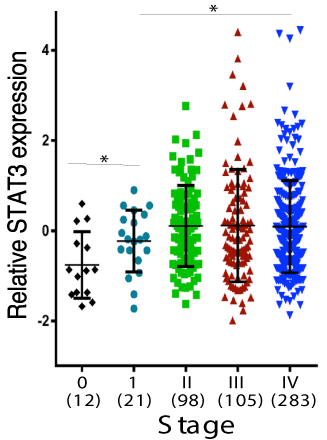
Figure 4. TCGA database. STAT3 expression in the TCGA database for head and neck cancers according to the Stage (Stage 0 is confined to the epithelium and Stage 4 with distant metastasis)
Gene Expression analysis of serially transplanted UTSCC 1 tumor tissue.
Since we found that the UTSCC-1 tumor tissue grew relatively slowly in vivo, we examined whether serial transplantation would improve tumor growth. While the original transplant of UTSCC-1 tumor tissue into the flanks of NSG mice took ~279 days to reach 1 cm in diameter, serially transplanted tumors only took 49 days to reach this size. Most interestingly, lung metastases were observed in mice with serially transplanted tumors. At necropsy we removed the tumor tissue and prepared RNA for gene expression analysis by qPCR. As shown in Figure 5, gene expression analysis of stem cell markers showed that SOX2 expression was slightly downregulated in serial transplanted tumor tissue, but its expression markedly downregulated in the lung metastasis tissue. In contrast, CD44 and Nestin was significantly upregulated in lung metastatic tissue. Expression of CD133 was markedly upregulated in serially transplanted tumor tissue, but not in lung metastasis.
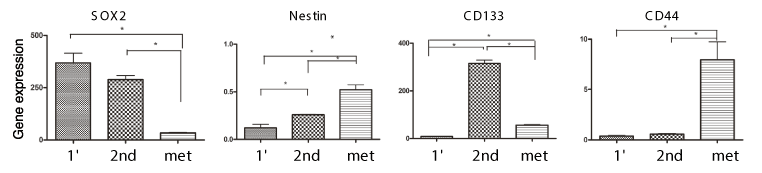
Figure 5. Gene Expression analysis of serially transplanted UTSCC-1 tumor tissue. Total RNA was prepared from the primary tumor transplant, serially transplanted tumor and lung metastatic tissue, and gene expression determined by qPCR, and normalized to actin expression
Furthermore, STAT3 expression was statistically significant increased in serially transplanted tumor tissue relative to expression in the original tumor (Figure 6). In addition, a statistical significant increase in STAT3 expression was found in lung metastasis as compared to the original tumor, as well as in the serially transplanted flank tumor.
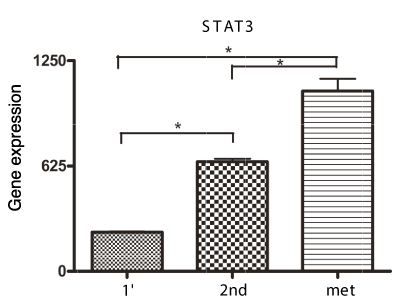
Figure 6. Gene Expression analysis of serially transplanted UTSCC-1 tumor tissue. Total RNA was prepared from the primary tumor transplant, serially transplanted tumor and lung metastatic tissue. STAT3 gene expression was determined by qPCR, and normalized to actin expression
We then investigated the expression of genes in the Notch pathway in these tumor tissues (Figure 7). NOTCH1, NOTCH3, and genes downstream in the Notch pathway such as JAG1, NUMBL, HES5 and DTX3 were overexpressed in lung metastasis tissue as compared to their expression in the primary and serially transplanted tumor tissue. NOTCH4 was significantly overexpressed in the serially transplanted tumor tissue and not in lung metastasis. However these alterations in Notch expression and downstream genes in the pathway were highly selective as there is no difference in the expression of NOTCH2 in primary or serially transplanted tumor tissue.
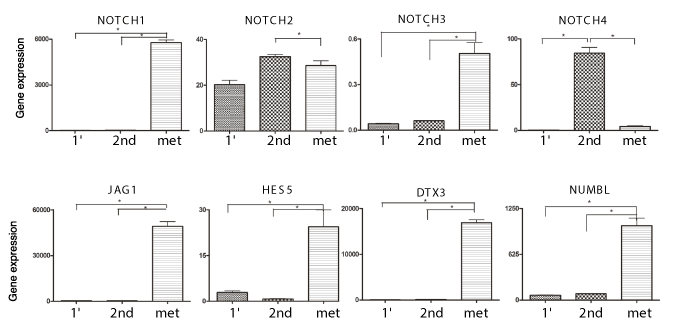
Figure 7. Gene Expression analysis of serially transplanted UTSCC-1 tumor tissue. Total RNA was prepared from the primary tumor transplant, serially transplanted tumor and lung metastatic tissue. Notch pathway gene expression was determined by qPCR, and normalized to actin expression.
Discussion
The survival rates in HNSCC have been relatively stagnant for decades with important risk factors being smoking use, alcohol use and HPV infection [1]. The overlap in these risk factors is believed to lead to the complex molecular alterations observed in HNSCC. In the current era of personalized therapy there is a promising role for selecting treatment according to the molecular alterations of tumors, such as in the case of lung cancer [20], but site and stage are still used for planning treatment in HNSCC [21]. The aim of the present study was to define the altered molecular pathways in HNSCC with the goal to eventually tailor treatment and overcome therapeutic resistance. We took the approach of establishing HNSCC patient-derived xenolines in immunocompromised nude mice because we found that HNSCC tumor tissue was very difficult to grow in tissue culture, which is consistent with the finding that only 3 out of 194 clinical samples could undergo more than 15 passages in vitro [22]. In addition the success rate of growing HNSCC tumor tissues in immunocompromised mice is relatively high, reportedly between 26% and 76% [23].
One important finding from our studies was that several CSCs markers, such as SOX2, CD44, and CD133, were overexpressed in one of the two HNSCC tumors studied, which appeared to be a less differentiated carcinoma by histological analysis. HNSCC cells that were CD44-positive were previously found to have tumorigenic potential in immunodeficient mouse models as compared to CD44-negative cells [24]. In addition, the CD133-positive population of laryngeal cancer cells was found to have a much higher tumorigenic potential in immunodeficient mice as compared to CD133-negative cells [25]. In more recent studies, SOX2 was identified as a CSC marker in HNSCC, and modulation of SOX2 expression by treatment with valproic acid can potently inhibit tumor growth and induce differentiation [26]. Our finding of enhanced CSC marker gene expression in HPV-HNSCC is clearly proof of the concept that CSCs play an important role in HNSCC tumorigenesis. In addition, although the sample number is small, the UTSCC-2 patient had high risk of smoking exposure and exhibited a statistically significant increase in the expression of CSC marker genes (CD44, CD133 and SOX2), which implies a possible role of smoking as a carcinogen in CSC enrichment. We previously enriched for glioma CSCs by growth in complete neural basal media on laminin-coated plates [12, 11]. In the present study we found that spheroid colonies of cells readily formed from UTSCC-2 tumor tissue under these conditions, but colonies did not form from UTSCC-1 tumor tissue or when UTSCC-2 tumor tissue was grown in serum-containing DMEM, which promotes cell differentiation. These results are further evidence of the enhanced CSC properties of UTSCC-2 tumors as compared to UTSCC-1 tumors. Moreover, serial transplantation of UTSCC-1 resulted in a significant upregulation of Nestin and CD133 expression, providing further evidence for a role of CSC marker genes in metastasis and tumor progression in HNSCC.
We have previously demonstrated STAT3 was constitutively activated in glioma CSCs, which resulted in enhanced expression of Notch pathway genes [12]. Another important finding in our study is that STAT3 gene expression was greater in the UTSCC-2 tumor that is more CSC-like as compared to UTSCC-1. The increased STAT3 expression was also confirmed, as well as that of the activated form of STAT3, by immunochemistry. STAT3 is constitutively activated in many cancers, including HNSCC and therefore has been targeted in clinical trials in HNSCC [27]. Furthermore, in a phase 0 clinical trial in HNSCC with a STAT3 decoy oligonucleotide, STAT3 was decreased in tumor tissue after decoy treatment [28]. Previous analysis also demonstrated a link between activated STAT3 (pSTAT3) and CSCs in HNSCC. For example a pharmacological STAT3 inhibitor eradicated the CSC population in a HNSCC cell line in vitro as well as tumor formation in vivo [29]. In the present study we found that serial transplantation of the UTSCC-1 tumor not only enhanced tumor growth and resulted in the formation of lung metastases, but also resulted in a statistically significant increase in STAT3 expression, which provides further evidence of an important role of STAT3 in HNSCC tumorigenesis and metastasis. In addition, by TCGA database analysis of a fairly large number of HNSCC tumor samples we found a statistically significant increase in the expression of STAT3 in higher stage tumors. Taken together these findings strongly implicate a critical role of STAT3 in HNSCC tumor progression.
Similar to normal stem cells CSCs have the capacity to self-renew and differentiate into multiple cell types, possibly mediating treatment resistance leading to recurrence or metastasis. Therefore, it is not surprising that critical signaling pathways, such as the Wnt, Notch and Hedgehog pathways, have been found to play important roles in CSCs and normal stem cells [30]. Another important finding in the present study is that Notch pathway genes are overexpressed in UTSCC-2 tumor tissue, implicating that this pathway is a potential drug target in HPV-HNSCC. In a comparative study of HNSCC tumor versus the adjacent normal tissue, there was distinct increase in the copy numbers in JAG1/JAG2, NOTCH1/3 and HES5 in tumor tissue [31]. It is interesting that we found that serially transplanted UTSCC-1 tissue showed a significant increase in the expression of several Notch pathway genes in lung metastatic tissue, implying its role in HNSCC metastasis.
HPV infection associated with HNSCC is becoming a major problem as it continues to rise. There is special need to understand the molecular biology of the disease. In this study we have shown that CSCs, and the STAT3 and Notch pathways appear to play an important role in HNSCC tumorigenesis and metastasis. Further studies are required to define the exact relationship between tobacco use and the CSC-like properties of HPV-HNSCC, the expression of STAT3 and that of Notch pathway genes. These findings open a new avenue to explore in HNSCC cancer research through a better understanding of the complex interactions between the causative factors leading to carcinogenesis and subsequent molecular pathway alterations that would allow for identification of new pharmaceutical targets.
Author contributions
Vijay G. Linga, Jessica Moskovitz, Debolina Ganguly, Michelle Sims, Louisa Balasz, Sandeep Samant and Lawrence M. Pfeffer conceived and designed the experiments; Vijay G. Linga, Debolina Ganguly, Michelle Sims, Andrew Dudas, Lindsey Cobb, Chuan He Yang and Louisa Balasz performed the experiments; Vijay G. Linga, Jessica Moskovitz, Debolina Ganguly, Michelle Sims, Chuan He Yang, Louisa Balasz and Lawrence M. Pfeffer analyzed the data; Jessica Moskovitz, Debolina Ganguly, Michelle Sims, Chuan He Yang, Louisa Balasz, Sandeep Samant and Lawrence M. Pfeffer contributed reagents/materials/analysis tools; Vijay G. Linga, Jessica Moskovitz, Debolina Ganguly, Sandeep Samant and Lawrence M. Pfeffer wrote the paper.
Competing interest
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by a West Cancer Center grant and funds from the Muirhead Chair Endowment at the University of Tennessee Health Science Center.
References
- Bhaijee F, Pepper DJ, Pitman KT, Bell D (2012) Cancer stem cells in head and neck squamous cell carcinoma: a review of current knowledge and future applications. Head Neck 34: 894-899.[Crossref]
- Suh Y, Amelio I, Guerrero Urbano T, Tavassoli M (2014) Clinical update on cancer: molecular oncology of head and neck cancer. Cell Death Dis 5: e1018. [Crossref]
- Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, et al. (2010) Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 363: 24-35. [Crossref]
- Kostareli E, Holzinger D, Hess J (2012) New Concepts for Translational Head and Neck Oncology: Lessons from HPV-Related Oropharyngeal Squamous Cell Carcinomas. Front Oncol 2: 36.[Crossref]
- Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, et al. (2011) Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 29: 4294-4301. [Crossref]
- Sayed SI, Dwivedi RC, Katna R, Garg A, Pathak KA, et al. (2011) Implications of understanding cancer stem cell (CSC) biology in head and neck squamous cell cancer. Oral Oncol 47: 237-243.[Crossref]
- Davis SJ, Divi V, Owen JH, Bradford CR, Carey TE, et al. (2010) Metastatic potential of cancer stem cells in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg 136: 1260-1266. [Crossref]
- Chinn SB, Darr OA, Owen JH, Bellile E, McHugh JB, et al. (2015) Cancer stem cells: mediators of tumorigenesis and metastasis in head and neck squamous cell carcinoma. Head Neck 37: 317-326. [Crossref]
- Shah A, Patel S, Pathak J, Swain N, Kumar S (2014) The evolving concepts of cancer stem cells in head and neck squamous cell carcinoma. Scientific World Journal 2014: 842491. [Crossref]
- Yadav A, Kumar B, Datta J, Teknos TN, Kumar P (2011) IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol Cancer Res 9:1658-1667. [Crossref]
- Garner JM, Ellison DW, Finkelstein D, Ganguly D, Du Z, et al. (2015) Molecular heterogeneity in a patient-derived glioblastoma xenoline is regulated by different cancer stem cell populations. PLoS One 10: e0125838. [Crossref]
- Garner JM, Fan M, Yang CH, Du Z, Sims M, et al. (2013) Constitutive Activation of Signal Transducer and Activator of Transcription 3 (STAT3) and Nuclear Factor kappaB Signaling in Glioblastoma Cancer Stem Cells Regulates the Notch Pathway. J Biol Che 288: 26167-26176. [Crossref]
- Yang CH, Shi W, Basu L, Murti A, Constantinescu SN, et al. (1996) Direct association of STAT3 with the IFNAR-1 chain of the human type I interferon receptor. J Biol Chem 271: 8057-8061.[Crossref]
- Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, et al. (1994) Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 77: 63-71. [Crossref]
- Zhong Z, Wen Z, Darnell JE Jr (1994) Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264: 95-98. [Crossref]
- Grandis JR, Drenning SD, Zeng Q, Watkins SC, Melhem MF, et al. (2000) Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc Natl Acad Sci U S A 97: 4227-4232. [Crossref]
- Poehlmann TG, Fitzgerald JS, Meissner A, Wengenmayer T, Schleussner E, et al. (2005) Trophoblast invasion: tuning through LIF, signalling via Stat3. Placenta 26 Suppl A: S37-41. [Crossref]
- Lino MM, Merlo A, Boulay JL (2010) Notch signaling in glioblastoma: a developmental drug target? BMC Med 8: 72. [Crossref]
- Deeken JF, Newkirk K, Harter KW, Marshall MB, Banovac F, et al. (2015) Effect of multimodality treatment on overall survival for patients with metastatic or recurrent HPV-positive head and neck squamous cell carcinoma. Head Neck 37: 630-635. [Crossref]
- Cagle PT, Raparia K, Portier BP3 (2016) Emerging Biomarkers in Personalized Therapy of Lung Cancer. Adv Exp Med Biol 890: 25-36. [Crossref]
- Mes SW, Leemans CR, Brakenhoff RH (2016) Applications of molecular diagnostics for personalized treatment of head and neck cancer: state of the art. Expert Rev Mol Diagn 16: 205-221.[Crossref]
- Krause CJ, Carey TE, Ott RW, Hurbis C, McClatchey KD, et al. (1981) Human squamous cell carcinoma. Establishment and characterization of new permanent cell lines. Arch Otolaryngol 107: 703-710. [Crossref]
- Baker SR (1985) An in vivo model for squamous cell carcinoma of the head and neck. Laryngoscope 95: 43-56. [Crossref]
- Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, et al. (2007) Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A 104: 973-978. [Crossref]
- Wei XD, Zhou L, Cheng L, Tian J, Jiang JJ, et al. (2009) In vivo investigation of CD133 as a putative marker of cancer stem cells in Hep-2 cell line. Head Neck 31: 94-101. [Crossref]
- Lee SH, Nam HJ, Kang HJ, Samuels TL, Johnston N, et al. (2015) Valproic acid suppresses the self-renewal and proliferation of head and neck cancer stem cells. Oncol Rep 34: 2065-2071.[Crossref]
- Song JI, Grandis JR (2000) STAT signaling in head and neck cancer. Oncogene 19: 2489-2495. [Crossref]
- Sen M, Thomas SM, Kim S, Yeh JI, Ferris RL, et al. (2012) First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer Discov 2: 694-705. [Crossref]
- Bu LL,, Zhao ZL, Liu JF, Ma SR, et al. (2015) STAT3 blockade enhances the efficacy of conventional chemotherapeutic agents by eradicating head neck stemloid cancer cell. Oncotarget 6: 41944-41958. [Crossref]
- Abetov D, Mustapova Z, Saliev T, Bulanin D, Batyrbekov K, et al. (2015) Novel Small Molecule Inhibitors of Cancer Stem Cell Signaling Pathways. Stem Cell Rev 11: 909-918. [Crossref]
- Sun W, Gaykalova DA, Ochs MF, Mambo E, Arnaoutakis D, et al. (2014) Activation of the NOTCH pathway in head and neck cancer. Cancer Res 74: 1091-1104. [Crossref]







