Induced pluripotent stem cells (iPSCs) bear potential to change the paradigms of conventional medicine. However, producing sufficient numbers of pluripotent and homogenous iPSC for therapeutic applications is challenging, in particular when using standard adherent cell culture. Suspension systems are prone to automation and thus represent a suitable alternative. Moreover, several well-established suspension culture vessels are already available. To support decision-making when establishing a cell production process, different culture systems were compared regarding the ability to standardize, product quality, economic efficiency, and potential for process optimization. Standard adherent culture was considered as a reference. In all experiments, the previously published AR1034ZIMA human iPSC line (ARiPS) was used. Culture systems comprised simple devices such as Petri dishes, Erlenmeyer flaks, as well as a computationally modelled and optimized stirred bioreactor. Highest standardization and cell yield was achieved when using the stirred bioreactor system, whereas Erlenmeyer flasks provided a good economic efficiency. Cell quality was comparable for all investigated vessels. Results indicate that the culture system should be carefully selected according to the application-specific requirements. Further, we show that low-cost culture vessels facilitate the production of high cell yields exhibiting a reasonable cell quality.
stirred bioreactor, induced pluripotent stem cells, stem cell culture, adherent and suspension stem cell culture, scale up, computational fluid dynamics, rheological fluid properties
Stem cells exhibit a high potential in basic research, e.g. to understand diseases, in drug discovery, as well as in cell-based therapies. Due to their theoretically unlimited self-sustainability and differentiation capacity, stem cells may allow to change the paradigms of conventional medicine by pharmacologically reinforcing aged and diseased tissues. Currently, therapies for the treatment of diseases like type 1 diabetes, Parkinson‘s disease, spinal cord injury, heart disease, and blindness are under investigation [1].
Induced pluripotent stem cells (iPSCs) are reprogrammed from mature somatic cells and allow insight into tissue regeneration and development. iPSCs can be differentiated into any mature cell type of the three embryonic germ layers [2], and facilitate in vitro disease models by utilizing patient-specific cells [3]. Diseased phenotypes are selectively generated, or cells with genetic defects may be reprogrammed with the aim to develop personalized therapies as well as novel drugs [4]. Although already tested in first clinical studies [5,6], producing feasible numbers of pluripotent and homogenous cell populations for Tissue Engineering and Regeneration remains a challenging task.
For cell-based therapeutic applications, 1×107 to 1×109 cells are required [7-10]. Afore mentioned cell numbers are beyond the capacity of adherent cell culture. T-flasks and multi-well plates, which are widely used in iPSC culture, entail extensive labor costs by binding experienced personnel [11]. For strictly adherent cultures, the adhesion surface area is commonly expanded by the use of micro-carriers in spinner flasks. However, recent experiments suggest that the fluid flow regime needs to be carefully characterized in order to avoid redundant physical stress since mechanical stimulation entails lineage commitment, and thus a loss of differentiation capacity [12,13]. Still, suspension cultures represent a considerable alternative culture strategy. Without micro-carriers, iPSCs are propagated as floating aggregates [14-17]. In contrast to standard adherent cell culture, suspension systems provide cost-efficient cell production [18]. However, transferring a specific iPSC line from standard adherent culture to suspension culture is challenging [19,20]. From an economic point of view, automated high-end suspension culture systems allow for reduced labor costs whereas intensive financial investment is required.
Currently, suspension culture strategies of various working volumes, manual operation and complexity are available. Here, Petri dishes placed on an orbital shaker are considered as the least complex culturing system. Under these conditions, Petri dishes are characterized by a chaotic flow regime, limitations regarding monitoring of process parameters, and comparably small working volumes. An increased working volume is feasible by using Erlenmeyer flasks. Like Petri dishes, the chaotic flow regime in an Erlenmeyer flask on a shaker can hardly be standardized, resulting in non-reproducible mechanical stimulation of cultured cells. In contrast to Petri dishes, online monitoring is possible. Commonly, single use sensors are therefore introduced into the culture vessel [21]. To avoid uncertainties due to undefined flow conditions, controlled mixing in stirred bioreactors is harnessed. Stirred bioreactors cover a broad range of working volumes up to 10.000 liters, and represent the gold standard in many biotechnological applications [22]. For the culture of mammalian cells, a variety of automated devices is commercially available. In addition to several online monitoring options, impellers exhibiting optimized geometries ensure defined agitation to support standardization [23]. However, high levels of defined and reproducible culture conditions in combination with process monitoring entail an increased technical complexity. A major advantage of controllable flow conditions in a stirred bioreactor system is the possibility to characterize the flow regime employing computational modelling. Thereby, mechanical stimuli such as shear stress are investigated, facilitating the identification of impeller rotational speeds that maintain cell aggregates afloat while minimizing mechanically-induced stress levels. Furthermore, computational model-derived mechanical culture conditions bear potential to guide cell fate [12].
In our study, we compared different culture systems for their ability to be standardized, product quality, economic efficiency, and potential for process optimization to support decision-making when designing human iPSC (hiPSC)-based production processes (Figure 1A). Characterized culture devices comprised agitated Petri dishes and Erlenmeyer flasks, and a computational fluid dynamics (CFD)-optimized stirred bioreactor. Adherent cell culture in well plates was used as a reference.
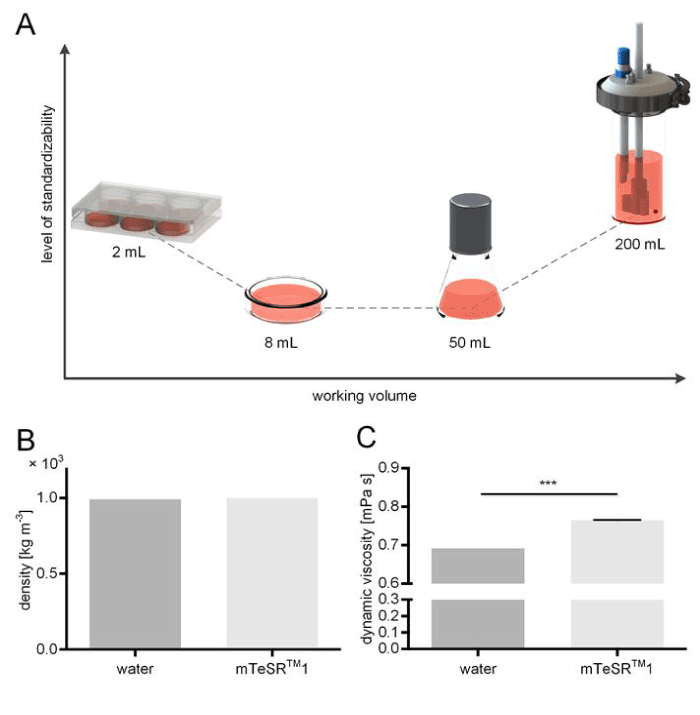
Figure 1.Schematic overview of culture vessels and fluid rheology measurements. (A) Suspended human induced pluripotent stem cells (hiPSCs) were cultured in medium volumes ranging from 8 mL (Petri dish), 50 mL (Erlenmeyer shaker flask), and at 200 mL scale in a custom made stirred tank bioreactor. In addition, adherent hiPSCs cultures were grown in standard 6-well plates as reference culture. (B) Comparison of fluid densities of water and stem cell culture medium mTeSRTM1 indicated no significant change among the measured fluids. (C) Values for the dynamic viscosity of water and mTeSRTM1 medium, respectively, were detected to differ significantly (n=3, p<0.05).
Adherent culture of human induced pluripotent stem cells
For our studies, we explicitly used hiPS cells of the ARiPS line. This cell line was generated from human adult fibroblasts using lentiviral vectors and subsequent TAT-Cre treatment for the removal of transgenes [24]. hiPSCs were maintained on hESC-qualified Matrigel®-coated (BD Biosciences #354277, Germany) 6-well plates (Nunc #140675) in mTeSRTM1 medium (STEMCELL Technologies #05850, Germany).
Cells were seeded at densities of 2×104 cells cm-2 for 6 days adherent culture and treated with 10 µM Y27632 ROCK-inhibitor (Sigma-Aldrich #Y0503-1MG, Germany) during the first 24 hours. Cell culture medium was replaced on a daily basis, and cell culture was performed in standard incubators at 37°C and 95% relative humidity and 5% CO2.
Suspension culture of human induced pluripotent stem cells
For suspension cultures, ARiPS were seeded at 1×105 cells mL-1 in suspension vessels and culturing was done for 6 days. Following, cells were treated with 10 µM Y27632 ROCK-inhibitor (Sigma-Aldrich) for 24 hours. To assess culture systems, hiPSC aggregate formation was compared for: (I) Petri dish culture, (II) Erlenmeyer flask, and (III) an in-house constructed stirred bioreactor. Petri dish and Erlenmeyer flask cultures were performed in standard incubators at 37°C, 95% relative humidity, and 5% CO2 on orbital shakers (Edmund Bühler GmbH, Germany), whereas hiPSC bioreactor cultures were performed in a tailor-made incubator at 37°C and 5% CO2. Where applicable, gas exchange was allowed by lid in place, comparable to a well plate. Gas exchange for the bioreactor was provided by two air filters (Sartorius Stedim Biotech GmbH, Germany). An overview of the different culture conditions and reaction volumes, rotational speeds and the initial hiPSC seeding densities is provided in table 1.
Table 1. Overview of the culture approaches of hiPSCs
| |
Culture volume |
Rotational speed [min-1] |
hiPSC inoculum |
| Adherent culture |
2 mL per well |
static |
2×104 cells cm-2 |
| Petri dish |
8 mL |
75 (orbital shaker) |
8×105 cells |
| Erlenmeyer flask |
50 mL |
120 (orbital shaker) |
5×106 cells |
| Bioreactor |
200 mL |
60 (electric drive) |
2×107 cells |
For ROCK-inhibitor removal, cell culture medium was replaced 24 hours after beginning the suspension culture. Following, 7/8 of the cell culture medium was replaced every 48 hours.
In vitro differentiation of human induced pluripotent stem cells
hiPSC sampled from culture vessels were seeded on 0.1% gelatine-coated (Sigma #G1890, Germany) 6-well plates. Cells were treated with mTeSRTM1 medium until cells reached 80 to 90% confluence. Unspecific differentiation was induced by DMEM (Gibco #41966-029, Germany) containing 10% fetal calve serum (Biochrom #S0115, Germany), 1% non-essential amino acids (Gibco #G11140-035, Germany), 1% sodium pyruvate (Gibco #1136-070, Germany), 1% L-glutamine (Gibco #25030-024, Germany), and 0.2% beta-mercaptoethanol (Gibco #31350-016, Germany). Differentiation was performed for 14 days and medium was replaced routinely every other day.
Stirred bioreactor
A custom-made stirred bioreactor was constructed from a glass vessel (Weckert, Germany), and a three-bladed impeller (Bohlender, Germany). The vessel was equipped with an ion-sensitive field-effect transistor (ISFET) pH sensor (Endress + Hauser GmbH, Germany) in addition to an optical oxygen probe (Presens, Germany). Mixing was achieved by connecting the agitator shaft to an electric drive (Faulhaber GmbH, Germany).
Fluid dynamics and shear stress estimation
To investigate the shear stress caused by agitation, a CFD model was established (COMSOL Multiphysics GmbH, Germany). Therefore, a three-dimensional (3D) to scale model of the fluid body was generated using computer-aided design (CAD) software (SolidWorks, Dassault Systemes, Germany) and imported into COMSOL. The model was composed of a rotating domain enclosing the impeller, and a static domain that portrayed the shape of the submerged pH probe. The two domains were linked via flow continuity boundary condition. An open boundary condition was defined at the air-liquid interface. Additionally, a pressure point constraint was defined to set ambient pressure conditions at the fluid surface. Gravity effects were considered by defining a volume force of according mass acceleration. For the subsequent numerical simulations, COMSOL’s rotating machinery module coupled to a κ-Ε turbulent flow model based on Reynolds-averaged Navier–Stokes equations was employed (CΕ1=1.44, CΕ2=1.92, Cµ=0.09, σκ=1.0, σΕ=1.3, κν=0.41, B=5.2). Values derived from density and dynamic viscosity measurements of mTeSRTM1 medium were used to adjust material properties. All remaining parameters were considered identical to those of water at 37°C.
Rheology
To enhance the precision and thus the prediction power of the CFD model, density and dynamic viscosity measurements of mTeSRTM1 cell culture medium were performed. Measurements were carried out using a density meter DMA4100M (Anton-Paar GmbH, Germany) and viscosity meter Lovis 2000M (Anton-Paar GmbH, Germany), respectively, following the manufacturer’s suggestions.
In silico particle sedimentation modelling and model validation
For modelling cell aggregate suspension, gelatine microspheres (Cultispher®, Sigma Aldrich, Seelze, Germany) of varying diameters (50-300 µm) were used as they showed comparable sedimentation behaviour to iPSC aggregates. In silico particles were generated employing COMSOL’s particle tracing module and their intrinsic geometrical attributes were adjusted to match the gelatine microspheres. In silico particles were implemented into the established CFD model and coupled to the stationary solution of the fluid velocity field. The fluid drag force was set to drive particle settling or elevation, respectively. A particle inlet was defined for the bottom boundary of the vessel and time dependent computations for particle elevation were performed for increasing impeller speeds. Particles that remained on the model’s bottom boundary after 3 seconds were counted and correlated to the corresponding impeller speed. For model validation, microspheres were homogenously distributed in the bioreactor without mixing until all microspheres had settled at the bottom of the vessel. Impeller-driven suspension of microspheres was visually observed for increasing agitation rates. Throughout validation experiments, the fluid was kept at 37°C to avoid changes of fluid density and viscosity.
Immunocytochemistry
Cells were washed with phosphate buffered saline (PBS, ThermoFisher Scientific #20012-019, Germany) and fixated with 4 % paraformaldehyde (Carl Roth #P087.3) for 10 minutes. If not noted separately, all steps for immunocytochemistry were performed at room temperature on a rocking shaker. Following an additional washing step with PBS (ThermoFisher Scientific), permeabilization and blocking for 1 hour with 0.1% TritonTM X-100 (Sigma-Aldrich #T8787-100ML, Germany) and 5 % donkey serum (Merck Millipore #S30-100ML, Germany) in PBS was performed. Primary antibody incubation was done for 1 hour in blocking solution. All antibodies are listed in table 2. Cells were washed 3 times for 5 minutes with PBS containing 0.5% TWEEN® 20 (Sigma-Aldrich #P7949-500ML, Germany). Corresponding secondary antibodies were diluted in antibody dilution solution (DCS #ALI20R500, Germany) and incubated for 1 hour at room temperature. After 3 additional washing steps of 5 minutes each, cells were covered with Fluoromount-GTM with DAPI (eBioscience #00-4959-52, Germany). Fluorescence images were acquired using BZ-9000 BIOREVO System (KEYENCE, Germany).
Table 2. Antibodies used for immunocytochemistry
| Antibody |
Species |
Dilution |
Order information |
| Alpha-1-Fetoprotein (AFP) |
rabbit |
1:400 |
Dako #A0008, Germany |
| Actin-Smooth Muscle (SMA) |
rabbit |
1:600 |
Spring Bioscience Corporation #E2460, USA |
| Tubulin β 3 (TUBB3) |
mouse |
1:5000 |
BioLegend #801201, UK |
| anti-mouse,Alexa Fluor 647 |
donkey |
1:1000 |
Invitrogen #A31571, Germany |
| anti-rabbit, Alexa Fluor 488 |
donkey |
1:1000 |
Invitrogen #A21206, Germany |
Flow cytometry analysis of stem cells
Flow cytometry was performed on single cells, treated with Accutase (Sigma-Aldrich #A6S64-100ML, Germany). If not noted separately, all steps for flow cytometry were performed at 4°C, centrifugation between washing steps and before replacing all solutions was done at 250 g for 5 minutes. Antibodies used for immunofluorescence stainings are listed in Table 3.
Table 3. Antibodies used for flow cytometry
| Antibody |
Species |
Dilution |
Ordering informations |
| SSEA4 – PE |
mouse |
1:25 |
BD #560128, Germany |
| SOX2 – FITC |
mouse |
1:25 |
eBioscience #53-9811-82, Germany |
| NANOG |
rabbit |
1:25 |
Abcam #ab80892, Germany |
| anti-rabbit Alexa Fluor 488 |
donkey |
1:100 |
Invitrogen #A21206, Germany |
Briefly, cells were transferred in centrifugation tubes and washed with FACS buffer composed of 1% fetal calve serum (Bio & SELL #ADD.0500, Germany) in PBS supplemented with 2 mM EDTA (Sigma-Aldrich #E5134-1KG, Germany). To stain cell surface antigens, 1×105 cells were diluted in 80 µL FACS buffer containing 1 µL FcR blocking reagent (MACS Miltenyi Biotec, Germany). Blocking was maintained for 20 minutes. Antibodies were diluted in FACS buffer and incubated for 30 minutes.
To stain intracellular transcription factors, cells were fixated and permeabilized with 250 µL BD Cytofix/CytopermTM solution (BD #554722, Germany) for 20 minutes. When using other than directly-labeled fluorescence antibodies, cells were washed 2 times with Perm/WashTM buffer (BD #554723, Germany) and blocked for 30 minutes with 5% donkey serum (Biozol Diagnostica #ECL-ECS0217D, Germany). For directly-labeled antibodies, blocking was omitted and antibodies were diluted in 1x Perm/WashTM buffer (BD).
After primary antibody incubation, cells were washed 3 times with FACS buffer (cell surface staining) or Perm/WashTM buffer (intracellular antigens). Secondary antibodies were diluted 1:100 and incubated for 30 minutes. Quantification of labeled cells was achieved by flow cytometer analysis (FACSCalibur, BD, Germany).
Quantitative real-time polymerase chain reaction
Prior to isolating RNA for quantitative real-time polymerase chain reaction (qRT-PCR) analysis, cells were separated with Accutase (Sigma-Aldrich, Germany). 5×105 hiPSCs were washed with PBS and transferred to a 1.5 mL reaction tube occupied with 350 µL RLT buffer + 1% β-Mercaptoethanol (Sigma Aldrich #M3148-25ML, Germany). RNeasy Micro Kit (Quiagen #74004) was used according to the manufacturer’s instructions. RNA content was determined by microplate reader Infinite M200 (TECAN). cDNA was generated from 0.5 µg RNA using iScriptTM cDNA Synthesis Kit (BIO-RAD #1708891) in a Thermocycler 48 (SensoQuest GmbH, Germany). qRT-PCR was performed with 1 µL cDNA dilution using the SsoFastTM EvaGreen® Supermix (BIO-RAD #1725202). Primers were designed with NCBI primer depot software (https://primerdepot.nci.nih.gov) and used at a concentration of 4 pmol µL-1. Sequences are listed in the table 4. PCR was performed using a SensQuest (BIO-RAD) thermocycler, where the annealing temperature was set to 60°C, and the elongation temperature to 72°C. Relative expression was quantified using the comparative cycle threshold (Ct) method, normalized to GAPDH and RPL4 expression. Fold difference calculation was done by ΔΔCt method.
Table 4. Primers used in qRT-PCR
Gene |
NCBI number |
Primer sequences |
GAPDH |
NM_002046 |
F: TGA CGC TGG GGC TGG CAT TG
R: GCT CTT GCT GGG GCT GGT GG |
RPL4 |
NM_000968 |
F: GCC TGC TGT ATT CAA GGC TC
R: GGT TGG TGC AAA CAT TCG GC |
SOX2 |
NM_003106 |
F: GCT TAG CCT CGT CGA TGA AC
R: AAC CCC AAG ATG CAC AAC TC |
NANOG |
NM_024865 |
F: ATG GAG GAG GGA AGA GGA GA
R: GAT TTG TGG GCC TGA AGA AA |
POU5F1 |
NM_203289 |
F: GGT TCT CGA TAC TGG TTC GC
R: GTG GAG GAA GCT GAC AAC AA |
Statistics
Quantitative data was analyzed for statistical differences using a one-way ANOVA employing Fisher’s least significant difference test. For all statistical tests a p-value <0.05 was considered significant, n=3 for all experiments.
In silico flow regime and shear stress assessment
The estimation of occurring shear stress is a major advantage when using CFD. Since the flow behaviour of any Newtonian fluid, and thus shearing potential, are determined by its corresponding density and dynamic viscosity [25], we investigated the intrinsic properties of mTeSRTM1 cell culture medium and considered the obtained values in the CFD model. A density of 9.93×102 kg m-3 was measured for mTeSRTM1 (Figure 1B). This density was equal to the density of water at 37°C [26]. In contrast, 0.765 ± 0.01 mPa s was measured for the dynamic viscosity of cell culture medium, which was significantly higher than the dynamic viscosity of 0.692 mPa s obtained for water (Figure 1C). The CFD model depicted in Figure 2A visualizes the fluid flow that was caused by the impeller. The streamlines and red arrows indicate the fluid’s flow path direction and magnitude, so that we can assume the medium to be principally accelerated in the lower region of the vessel. This is also where the fluid is calculated to reach its peak flow velocity. Once the medium hits the reactor wall, it is then redirected upwards along the flow barrier. Based on the CFD model, shear stress data was extracted when stirring at different rotational speeds. Therefore, in silico domain probes indicating the maximum shear stress within the model’s fluid domains were deployed. For the investigation, a range of possible impeller rotational speeds ranging from 20-100 revolutions per minute (RPM) was covered. In consequence, the obtained shear stress levels ranged from 4 mPa up to 12.5 mPa in a non-linear fashion (Figure 2B).

Figure 1.Schematic overview of culture vessels and fluid rheology measurements. (A) Suspended human induced pluripotent stem cells (hiPSCs) were cultured in medium volumes ranging from 8 mL (Petri dish), 50 mL (Erlenmeyer shaker flask), and at 200 mL scale in a custom made stirred tank bioreactor. In addition, adherent hiPSCs cultures were grown in standard 6-well plates as reference culture. (B) Comparison of fluid densities of water and stem cell culture medium mTeSRTM1 indicated no significant change among the measured fluids. (C) Values for the dynamic viscosity of water and mTeSRTM1 medium, respectively, were detected to differ significantly (n=3, p<0.05).
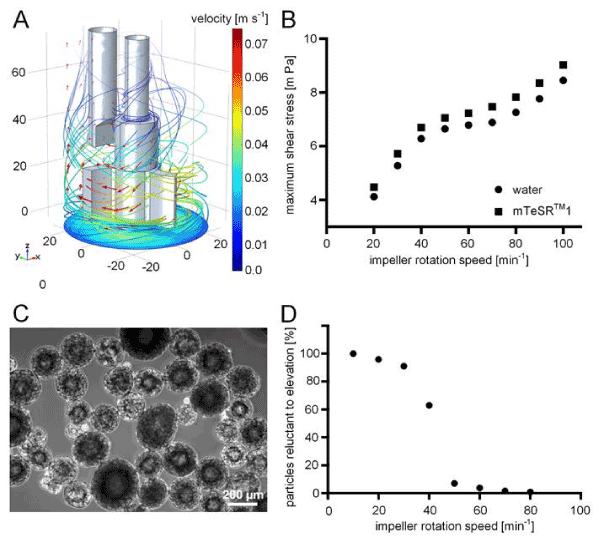
Figure 2.Implementation of computational fluid dynamics on the stirred tank bioreactor system. (A) Three dimensional (3D) computational model of the submerged stirrer and pH electrode inside the bioreactor. The depicted fluid velocity field (red arrows and streamlines) was caused by stirring at 30 revolutions per minute (RPM) and was obtained using finite element method (FEM) simulation software. The impeller geometry induced a radial flow field, which is guided upwards when the fluid hits the vessel jacket. The velocity magnitude is indicated as colour range. (B) Extracted data on maximum shear stress that was introduced into the fluid at distinct impeller rotational speeds. (C) Gelatine microspheres were used as model particles mimicking hiPSC aggregate sedimentation behaviour and were employed to identify the critical agitation rate at which particle suspension is ensured. (D) Percentage of suspended particles was simulated for an impeller rotational speed range of 10-80 RPM. Coupling of the CFD model with particle physics allowed the calculation of the critical rotation speed at which cell sedimentation is overcome. Continuous particle suspension was ensured at 50 RPM.
Particle sedimentation modelling
During suspension culture, it is commonly observed that hiPSCs form aggregates that are prone to sedimentation. By applying drag forces, for instance by mixing, sedimentation is delayed and, when surpassing a specific power input, sedimentation is prevented. In order to identify the critical impeller agitation rate that hampers aggregate sedimentation at the lowest shear stress input possible, we extended the existing CFD model by introducing in silico particles representing cellular aggregates. Therefore, we used COMSOL’s particle tracing module to define spheres of sizes and density comparable to that of cellular aggregates generally found in suspension cultures. A species of n=1000 particles with normally distributed diameters (mean: 0.2 mm, standard deviation: ± 0.05 mm) was generated at the bottom boundary of the computational model and particles were elevated by the drag force of the flow field caused by the moving impeller. Time-dependent studies of 3 seconds each were performed for a range of impeller speeds and the percentage of particles that remained on the model’s bottom boundary was correlated to the respective stirring speed (Figure 2D). Validation experiments revealed a high predictive power of the in silico model (Figure 3).
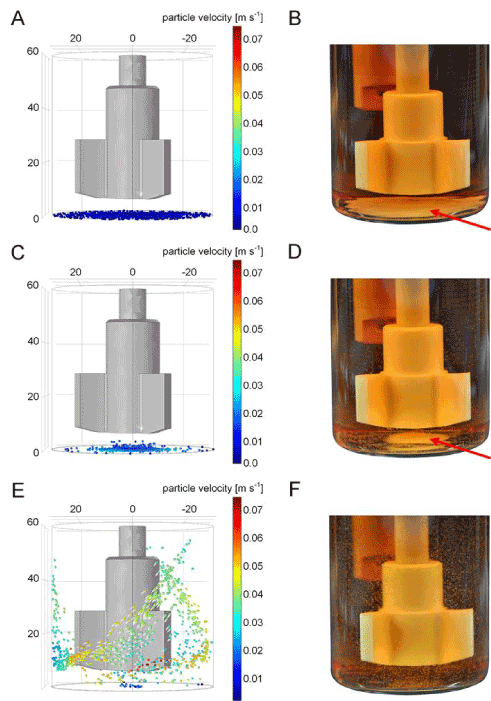
Figure 3.Validation of iPSC aggregate sedimentation modelling. A quantity of n=1000 in silico particles was generated at the bottom boundary of the vessel and particles were elevated by the impeller-derived flow velocity field. Simulation-based suspension capabilities are shown for (A) 10, (C) 30, and (E) 50 revolutions per minute. (B, D, F) Gelatine microsphere validation experiments at respective impeller speeds. Microsphere sedimentation (red arrows) is intensified when mixing below 50 RPM.
Process parameter monitoring
Medium conditions were monitored throughout the entire hiPSC suspension culture process for bioreactor experiments. Optical fibre probes were used to measure dissolved oxygen content in a non-invasive fashion. For pH value acquisition, continuous measurements were performed by using a conventional pH dipping probe. An overall decrease of both oxygen (Figure 4A) and pH (Figure 4B) was observed during cultivation. Medium exchange at indicated time points (dotted vertical lines) led to the replenishment of nutrients and dissolved oxygen. Aggregate diameter was qualitatively investigated during medium exchange steps.
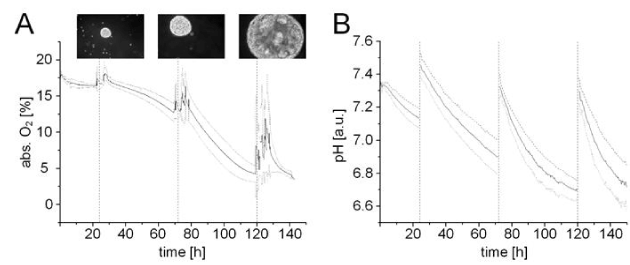
Figure 4.Online process monitoring. Visualization of acquired measurements for (A) oxygen and (B) pH levels. Between two medium exchanges (vertical slashed lines), oxygen consumption was observed to increase while pH levels decreased steadily due to growing cell numbers and, consequently, increasing hiPSC aggregate diameters (n=3), microscopic magnification 50 fold.
Suspension culture of hiPSCs
Without any possibility to adhere, hiPSCs are characterized by spontaneous aggregate formation. For the ARiPS cell line, hiPSC aggregates were formed during the first 24 hours in all suspension vessels. After 6 days, the diameter of the hiPSC aggregates from all vessels was similar with a mean of about 260 µm. All aggregates showed a homogenous, spherical morphology with an even contour. In contrast, adherent hiPSCs formed compact colonies with defined borders. The stirred bioreactor’s reaction volume of 200 mL resulted in a yield of up to 4.4×107 hiPSCs within 6 days (Figure 5A). For a comparison, 7 6-well plates of adherently cultured hiPSCs were required to obtain the same amount (cell yield of 1×105 hiPSCs cm-2). A single Petri dish and Erlenmeyer flask culture facilitated a yield of 5.8×106 cells and 1.48×107 hiPSCs, respectively.
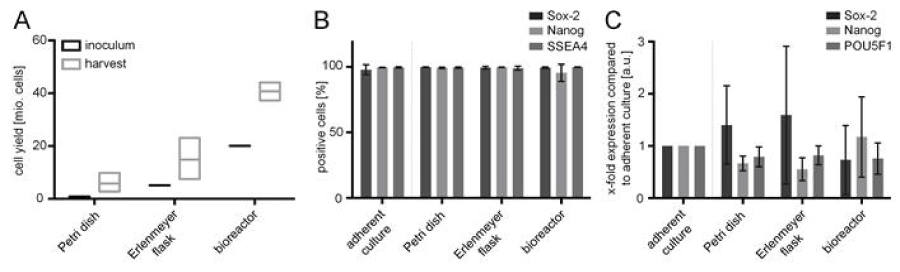
Figure 5.Cell yield after suspension culture and in vitro characterization of pluripotency. (A) Comparison of hiPSC yield between seeded cell number (inoculum) and after 6 days of culture. (B) Flow cytometry analysis of harvested hiPSCs in the different approaches. Cells were positive for the pluripotency-associated markers SOX2, NANOG, and SSEA4. Flow cytometry signals demonstrated consistent marker expression in all vessels. (C) qRT-PCR analyses of the transcription factors SOX2, NANOG and POU5F1 shown as change in gene expression (ΔΔCt) compared to the adherent standard culture. Measurements were normalized to housekeeping genes GAPDH and RPL4. All results are shown as mean ± SD (n=3), and each biological replicate represents an independent experiment.
Characterization of pluripotent stem cells after suspension culture in different culture vessels
In addition to cell morphology and hiPSC yield, pluripotency was investigated. Therefore, cells from all vessel types were characterized and compared on protein and gene level. In this context, hiPSCs were analyzed by flow cytometry for characteristic pluripotency-associated markers comprising transcription factors SOX2 and NANOG, and surface antigen SSEA4 (Figures 5B and 6). Flow cytometry revealed that at least 95% hiPSCs were found positive for all investigated pluripotency markers. Protein expression in cells cultured in suspension vessels and the adherent control showed no significant difference. Pluripotency of hiPSCs was confirmed on gene level by qRT-PCR analysis. The relative gene expression of transcription factors SOX2, NANOG and POU5F1 (also known as OCT3/4) was compared between adherent control (100%) and suspension cultures (Figure 5C). The variations in gene expression between hiPSCs cultured in the stirred bioreactor and hiPSC standard adherent cultures were lower than ± 27% and, consequently, had no biological relevance. The expression of NANOG, which is known to be distributed inhomogenously in pluripotent stem cell populations, was reduced by approximately 33 % in Petri dish, and by 45% in Erlenmeyer flask cultures. However, the expression of POU5F1 was slightly reduced in both culture vessels. Moreover, stem cells of stirred bioreactor cultures showed lower SOX2 expression compared to the adherent hiPSC control. In contrast, SOX2 expression was increased by 40 % after Petri dish cultivation and by 60% after Erlenmeyer flask culture. Finally, when using the in-house constructed stirred bioreactor, the expression of pluripotency-associated markers of hiPSCs was preserved.
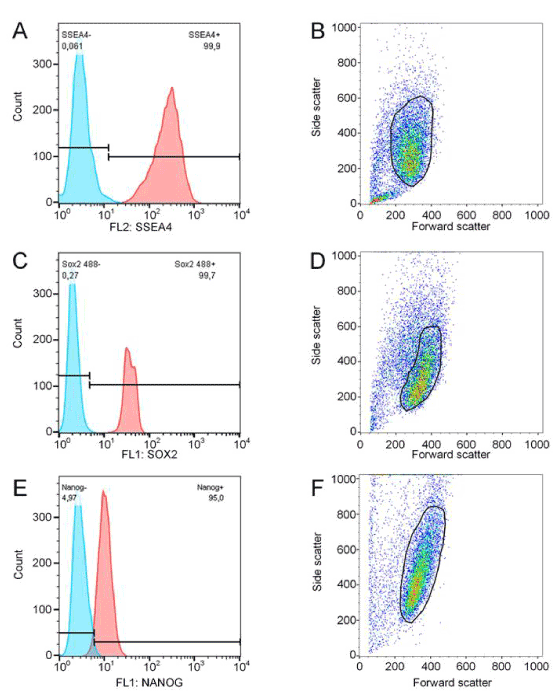
Figure 6.Flow cytometry analysis data to figure 5 B, exemplarily shown for one bioreactor experiment. Harvested cells were stained for SOX2 (A-B), NANOG (C-D), and SSEA4 (E-F). Cell populations were gated as indicated and subsequently analyzed for pluripotency marker expression.
Differentiation capacity of hiPSCs after stirred tank bioreactor culture
2021 Copyright OAT. All rights reserv
In order to confirm the pluripotency of hiPSCs after 6 days of culture in the stirred bioreactor, in vitro differentiation studies were performed and results were compared to adherent control cells. The capacity of both hiPSC groups to differentiate into cells of the endo-, meso- and ectoderm germ layers was demonstrated by immunofluorescence stainings. Here, the expression of characteristic markers Alpha-1-Fetoprotein (AFP; Figures 7A and 7B), smooth muscle actin (SMA; Figures 7C and 7D) and Tubulin β 3 (TUBB3; Figures 7E and 7F) was investigated. Interestingly, a variation in TUBB3 expression was detected. TUBB3 antibodies normally bind to cells with neuronal, elongated outgrowths. This was found true for bioreactor cultured cells (Figure 7F). Yet, TUBB3-positive differentiated cells that were derived from the adherent control culture (Figure 7E), were mainly not characterized by neurite-like processes, indicating an immature stage of neural progenitor cells.
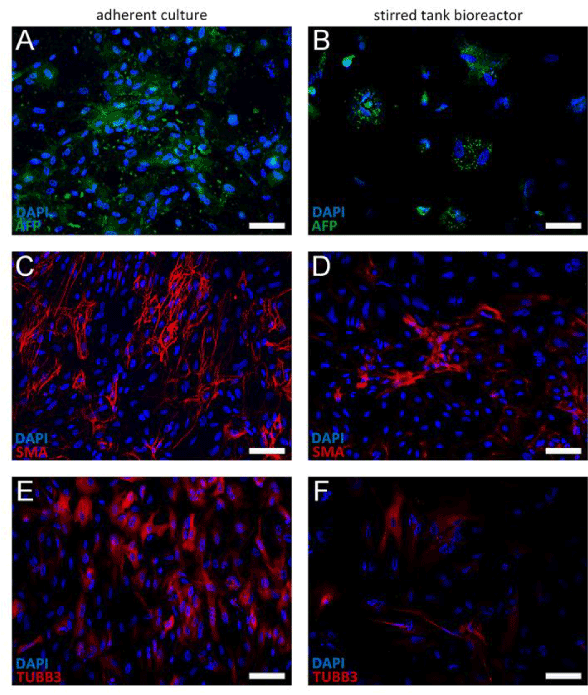
Figure 7.Immunohistochemistry analyses of differentiated hiPSCs by immunofluorescence staining of characteristic markers representative for the three germ layers. Differentiation potential of cells harvested from the stirred bioreactor was compared to the adherent culture control. Cells were stained for Alpha-1-Fetoprotein (AFP; A-B), smooth muscle actin (SMA; C-D) and Tubulin β 3 (TUBB3; E-F), demonstrating the capacity to differentiate into endoderm, mesoderm, and ectoderm germ layers. Cell nuclei were stained with DAPI in blue. Scale bars indicate 100 µm.
The scale-up of bioreactors for mammalian cell culture represents a technological challenge and requires reliable and efficient scaling methodologies [19]. In particular, the successful transfer of static adherent monolayer cultures to a 3D suspension environment is vital, since dynamic conditions show a considerable impact on cell behavior [27]. Due to the mechanical sensitivity of mammalian cells and the influence of shear stress, mechanical stimuli on cell fate have to be considered to ensure consistent product quality. In addition, homogenous oxygen and nutrient distribution must be controlled throughout the entire cell culture process. Thus, the way of introducing agitation and, consequently, the magnitude of induced mechanical stress should be carefully designed and implemented. Nevertheless, reproducible flow and standardized culture conditions entail a high degree of technical complexity, in particular when increasing the working volume for process scale up. In this context, we characterized different culture systems of varying working volumes for their capacity to be standardized (Figure 1A). Therefore, we cultured a hiPSC line, which exhibited a promising potential for therapeutic applications [24]. Based on the culture process results, the employed culture vessels were assessed regarding their ability to be standardized, yield and product quality, economic efficiency, as well as potential for process optimization.
Standardization
The highest level of standardization was provided by our stirred bioreactor system. In contrast to chaotically-agitated culture vessels such as Petri dish and Erlenmeyer flasks, the flow conditions inside the stirred bioreactor could be described in a reasonable approximation, and facilitated the development of a fluidic in silico model. Although a variety of computational models for stirred bioreactor systems have already been published [28,29], most models lack a realistic parametrization. For instance, rheological fluid properties are often considered to be identical to those of water. A comparison of the dynamic viscosity of cell culture medium, revealed significant variations, resulting in higher shear stress levels. To ensure a sufficient predictive precision, the model was successfully validated, and flow conditions were correlated to distinct impeller speeds. The chaotic flow regimes present in Petri dishes and Erlenmeyer flasks are challenging to validate [30]. In addition to defined flow conditions, the stirred bioreactor allowed for continuous monitoring of oxygen and pH to improve in-process monitoring. Systems exist that overcome carbon dioxide and oxygen variations during culture handling. Additionally, small scale culture can be performed at constant environmental conditions. Such cultivation devices offer a high level of standardization at considerable system complexity [31].
Yield and product quality
A successful scale-up increases cell yield while retaining consistent product quality [32]. When comparing cell yield, the stirred bioreactor facilitated the production of approximately 4×107 human iPSCs within 6 days of culture. The obtained cell numbers are in the lower range of those required for cell-based therapeutic applications [10]. Solely Erlenmeyer flask cultures reached comparable cell numbers. Nevertheless, stirred bioreactor cultures exhibited less variation in cell yield. A limitation of both systems is the high initial cell number that is required to start the culture process. Concerning the adherent culture process, the ratio between initial and final cell numbers was increased fivefold.
In comparison to the reference culture, all tested vessels exhibited consistent cell quality on both gene and protein level. Hereby, analyzed markers comprised the transcription factors NANOG and SOX2 as well as the membrane antigen SSEA4 on protein level. On gene level, the expression of NANOG, SOX2 and POU5F1 (Oct3/4) was confirmed. Despite the advantages of standard adherent hiPSC culture, such as gradient-free feeding of cells (monolayer vs. aggregate), and microscopic monitoring of morphology [33], we suggest a cascade bioreactor approach with increasing working volume. Thereby, low initial cell numbers are possible, and by transferring harvested cells to a greater working volume, large quantities of stem cells can be achieved. Nevertheless, consistent cell quality over several passages needs to be demonstrated.
Economic efficiency
When assuming an average cell number of 1×108 cells, which is within range of cell numbers required for therapeutic applications, approximately 18 6-well plates, 17 Petri dishes, 7 Erlenmeyer flasks, and 3 (2.5) stirred bioreactors are needed. This calculation is based on the cell yields obtained after 6 days of culture. While adherent hiPSC culture routinely requires a daily medium change, the medium of the suspension cultures was changed every other day. The lowest media consumption per culture process and vessel was required by Petri dish cultures (29 mL), followed by adherent cultures (72 mL), and Erlenmeyer cultures (181 mL). Bioreactor cultures consumed 725 mL mTeSRTM1 within 6 days. With regard to the amount of medium that is required to generate 1×108 cells, costs become relativized due to the number of required vessels. Stated in numbers, the total media consumption starting with the lowest is ranked as follows: 499 mL (Petri dish), 1202 mL (adherent culture), 1233 mL (Erlenmeyer flask) and 1668 mL (stirred bioreactor). In contrast to material costs, expenses for laboratory space and labor are less for systems with higher working volume. The total time and space required to process 17 Petri dishes is much greater compared to a single bioreactor where medium is exchanged every second day. In particular the standard adherent culture in 18 6-well plates, demanding for daily medium exchange, is disproportional in effort compared to Erlenmeyer flasks and stirred bioreactors.
Potential for process optimization
A major advantage of stirred bioreactors is the use of advanced probing devices such as sensors for dissolved oxygen and pH. This is mainly limited by the dimension-driven possibilities of the selected culture vessel and, in addition, by the method of agitation. Whereas it is comparably tedious to collect online measurement data from Petri dishes and Erlenmeyer flasks while placed on orbital shakers, sensors are easily connected to stirred tanks. The same holds when opting for oxygen and pH control in the medium. The required control appliances are in most cases already incorporated in commercially available stirred tank solutions [34-36]. Here, cell expansion under set and stable oxygen content and pH represents the major advantage of aforementioned systems, comprising hypoxic culture conditions if required.
Since stirred bioreactor agitation is prone to computational modelling, it is possible to obtain reasonable data on shear stress which can be easily correlated to the impeller speed (Figure 2B). However, while all of the above is merely feasible for more complex devices such as stirred tanks with suitable port inlets, oxygen and pH control in well plates, Petri dishes, and flasks is more elaborate and unsuitable for automation. Still, shake flask readers are available for measuring oxygen and pH in fluids while shaking [37]. On the other hand, the resulting increase of system complexity and material cost needs to be considered.
When designing a suspension culture process for iPSCs, the culture vessel should be carefully selected. Our study demonstrates that even simple vessels such as Erlenmeyer flasks allow for the expansion of iPSCs in a range that is required for therapeutic applications with acceptable effort. Hereby, sufficient cell quality comparable to adherent culture is possible. Nevertheless, controlled stirred bioreactor systems facilitate standardization, and thus the integration of a culture process into an automated environment.
Antje Appelt-Menzel: hIPSC culture, cell characterization, analysis of results, preparation of the manuscript; Ivo Schwedhelm: bioreactor development, computational modelling, bioreactor culture, analysis of results, preparation of the manuscript; Fabian Kühn: bioreactor development, hiPSC culture; Alevtina Cubukova: hiPSC culture, cell characterization; Frank Edenhofer: differentiation studies, preparation of the manuscript; Heike Walles: analysis of the results, preparation of the manuscript; Jan Hansmann: Study design, bioreactor development, analysis of the results, preparation of the manuscript.
Our work was funded by the German Federal Ministry of Education and Research; program NanoMatFutur; grant agreement number 13N12971 – ETface.
- Mimeault M, Hauke R, Batra S (2007) Stem cells: a revolution in therapeutics-recent advances in stem cell biology and their therapeutic applications in regenerative medicine and cancer therapies. Clin Pharmacol Ther 82: 252-264. [Crossref]
- Lewandowski J, Kurpisz M (2016) Techniques of Human Embryonic Stem Cell and Induced Pluripotent Stem Cell Derivation. Arch Immunol Ther Exp 1-22.
- Tzatzalos E, Abilez OJ, Shukla P, Wu JC (2016) Engineered heart tissues and induced pluripotent stem cells: Macro-and microstructures for disease modelling, drug screening, and translational studies. Advanced drug delivery reviews 96: 234-244.
- Grskovic M, Javaherian A, Strulovici B, Daley GQ (2011) Induced pluripotent stem cells-opportunities for disease modelling and drug discovery. Nat Rev Drug Discov 10: 915-929.
- Cyranoski D (2014) Japanese woman is first recipient of next-generation stem cells. Nature 10.
- Kamao H, Mandai M, Okamoto S, Sakai N, Suga A, et al. (2014) Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem cell reports 2: 205-218. [Crossref]
- Chen SL, Fang W-w, Ye F, Liu Y-H, Qian J, et al. (2004) Effect on left ventricular function of intracoronary transplantation of autologous bone marrow mesenchymal stem cell in patients with acute myocardial infarction. Am J Cardiol 94: 92-95. [Crossref]
- Connick P, Kolappan M, Patani R, Scott MA, Crawley C, et al. (2011) The mesenchymal stem cells in multiple sclerosis (MSCIMS) trial protocol and baseline cohort characteristics: an open-label pre-test: post-test study with blinded outcome assessments. Trials 12: 62. [Crossref]
- Jiang R, Han Z, Zhuo G, Qu X, Li X, et al. (2011) Transplantation of placenta-derived mesenchymal stem cells in type 2 diabetes: a pilot study. Front Med 5: 94-100. [Crossref]
- Le Blanc K, Frassoni F, Ball L, Locatelli F, Roelofs H, et al. (2008) Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. The Lancet 371: 1579-1586. [Crossref]
- Koller M, Emerson SG, Palsson B (1993) Large-scale expansion of human stem and progenitor cells from bone marrow mononuclear cells in continuous perfusion cultures. Blood 82: 378-384. [Crossref]
- Altman G, Horan R, Martin I, Farhadi J, Stark P, et al. (2002) Cell differentiation by mechanical stress. Faseb J 16: 270-272. [Crossref]
- Rozwadowska N, Malcher A, Baumann E, Kolanowski TJ, Rucinski M, et al. (2016) In vitro culture of primary human myoblasts by using the dextran microcarriers Cytodex3®. Folia Histochem Cytobiol. [Crossref]
- Cormier JT, Nieden NIZ, Rancourt DE, Kallos MS (2006) Expansion of undifferentiated murine embryonic stem cells as aggregates in suspension culture bioreactors. Tissue Eng 12: 3233-3245. [Crossref]
- Kempf H, Andree B, Zweigerdt R (2016) Large-scale production of human pluripotent stem cell derived cardiomyocytes. Adv Drug Deliv Rev 96: 18-30. [Crossref]
- Oh SK, Chen AK, Mok Y, Chen X, Lim U-M, et al. (2009) Long-term microcarrier suspension cultures of human embryonic stem cells. Stem cell research 2: 219-230. [Crossref]
- Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW (2008) Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PloS one 3: e1565. [Crossref]
- Chen VC, Ye J, Shukla P, Hua G, Chen D, et al. (2015) Development of a scalable suspension culture for cardiac differentiation from human pluripotent stem cells. Stem Cell Res 15: 365-375. [Crossref]
- Godoy‐Silva R, Berdugo C, Chalmers JJ (2010) Aeration, mixing, and hydrodynamics, animal cell bioreactors. Encyclopedia of Industrial Biotechnology, John Wiley & Sons, USA.
- Griffiths JB (2010) Mammalian Cell Culture Reactors, Scale‐Up. Encyclopedia of Bioprocess Technology, John Wiley & Sons, USA.
- Schiefelbein S, Fröhlich A, John GT, Beutler F, Wittmann C, et al. (2013) Oxygen supply in disposable shake-flasks: prediction of oxygen transfer rate, oxygen saturation and maximum cell concentration during aerobic growth. Biotechnol Lett 35: 1223-1230. [Crossref]
- Chu L, Robinson DK (2001) Industrial choices for protein production by large-scale cell culture. Curr Opin Biotechnol 12: 180-187. [Crossref]
- Platas Barradas O, Jandt U, Phan M, Da L, Villanueva ME, et al. (2012) Evaluation of criteria for bioreactor comparison and operation standardization for mammalian cell culture. Engineering in Life Sciences 12: 518-528. 24.
- Kadari A, Lu M, Li M, Sekaran T, Thummer RP, et al. (2014) Excision of viral reprogramming cassettes by Cre protein transduction enables rapid, robust and efficient derivation of transgene-free human induced pluripotent stem cells. Stem Cell Res Ther 5: 1-10. [Crossref]
- Kell Schnittler HJ, Franke RP, Akbay U, Mrowietz C, Drenckhahn D (1993) Improved in vitro rheological system for studying the effect of fluid shear stress on cultured cells. Am J Physiol 265: C289-C298. [Crossref]
- Kell G (1967) Precise representation of volume properties of water at one atmosphere. J Chem Eng Data 12 :66-69.
- Karimi M, Bahrami S, Mirshekari H, Basri SM, Nik AB, et al. (2016) Microfluidic systems for stem cell-based neural tissue engineering. Lab Chip. [Crossref]
- Qi N, Zhang H, Zhang K, Xu G, Yang Y (2013) CFD simulation of particle suspension in a stirred tank. Particuology 11: 317-326.
- Tamburini A, Cipollina A, Micale G, Brucato A, Ciofalo M (2013) CFD simulations of dense solid–liquid suspensions in baffled stirred tanks: Prediction of solid particle distribution. Chemical engineering journal 223: 875-890.
- Mancilla E, Palacios-Morales C, Córdova-Aguilar M, Trujillo-Roldán M, Ascanio G, et al. (2015) A hydrodynamic description of the flow behavior in shaken flasks. Biochem Eng J 99: 61-66.
- Stover A, Herculian S, Banuelos MG, Navarro SL, Jenkins MP, et al. (2016) Culturing Human Pluripotent and Neural Stem Cells in an Enclosed Cell Culture System for Basic and Preclinical Research. J Vis Exp [Crossref]
- Soares FA, Chandra A, Thomas RJ, Pedersen RA, Vallier L, et al. (2014) Investigating the feasibility of scale up and automation of human induced pluripotent stem cells cultured in aggregates in feeder free conditions. J Biotechnol 173: 53-58. [Crossref]
- Mummery CL, Zhang J, Ng ES, Elliott DA, Elefanty AG, et al. (2012) Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes a methods overview. Circ Res 111: 344-358. [Crossref]
- Ellert A, Vikström C (2014) Design of Experiments with Small-Scale Bioreactor Systems. Innovations in Cell Culture: 10.
- Olmer R, Kropp C, Huether-Franken C, Zweigerdt R (2013) Scalable expansion of human pluripotent stem cells in Eppendorf BioBLU® 0.3 single-use bioreactors. Eppendorf, Hamburg.
- Weber W, Rimann M, de Glutz F-N, Weber E, Memmert K, et al. (2005) Gas-inducible product gene expression in bioreactors. Metab Eng 7: 174-181. [Crossref]
- Lavrentieva A, Majore I, Kasper C, Hass R (2010) Effects of hypoxic culture conditions on umbilical cord-derived human mesenchymal stem cells. Cell Commun Signal 8: 1. [Crossref]







