Objectives: To examine the incidence and clinical dynamics of IKZF1 alterations in Chinese adult patients with ALL.
Methods: Samples were studied from 328 newly diagnosed adult ALL patients at Peking University People’s Hospital from 2007 to 2012 by multiplex real-time quantitative PCR, multiplex fluorescent PCR and sequence analysis for four types of IKZF1 deletions. Albumin (ALB) was the plasmid standard.
Results: All correlation coefficients for amplified ALB(albumin) and IKZF1 Δ4-7 deletion plasmids were above 0.99, and the sensitivity of detection was at least ten copies. About 36.3% (95% [CI], 31.1-41.7%) of 328 untreated cases showed IKZF1-deletion. The median IKZF1-deletion copies/ALB copy was 85.8% (0.1%-697.9%). 27 IKZF1-deletion-positive patients (219 samples in total) were followed up after treatment, among them, 18 patients in hematologic remission continued to be tested negative within 8-66 weeks. In 5 relapsed cases, elevated levels of IKZF1 deletion were detected when relapse occurred. In 4 non responders, IKZF1 deletion maintained at high level. Patient with B-cell ALL subtype had much higher IKZF1-deletion rate (40.4%) than those with T-cell ALL subtype (2.8%, P<0.01). In particular, it was higher in common B-cell ALL group than those with other subtypes.
Conclusion: This assay is reliable and sensitive. It is useful in diagnosis and monitoring minimal residual disease in adult ALL. High frequency of IKZF1 deletions occur in adult patients with common B-cell ALL.
IKZF1 deletion, acute lymphoblastic leukemia, real-time quantitative PCR
Genetic prerequisites for developing acute lymphoblastic leukemia (ALL) have been characterized during the past decades. Based on cytogenetic and molecular analyses, ALL carries translocations t(9;22)/(BCR/ABL), t(12;21)/ETV6-RUNX1, t(1;19)/E2A-PBX1, rearrangements of MLL, hyperdiploid or hypodiploid karyotypes, most of which were found to be associated with distinct prognosis and are currently used for risk-adapted treatment protocols [1,2]. Although these oncogenic lesions are crucial in leukemia initiation, they are not sufficient to define the biology of leukemia [1,3,4]. Besides these classifying lesions, a number of co-operating genetic lesions have recently been identified by genome-wide approaches [5-7]. Of particular interest are IKAROS (IKZF1) alterations, found not only in most BCR/ABL positive ALL but also in a small fraction of BCR/ABL negative ALL [6-10]. We and others has proved that such alterations are associated with a poor prognosis [11-16] and may act as a promising diagnostic and prognostic marker.
IKZF1 deletions comprise whole gene deletions, that can be the result of monosomy 7 or del (7p), and intragenic deletions. Most IKZF1 intragenic deletions encompass exons 4 to7 (thereafter named Δ4-7), driving the expression of a non-DNA binding Ikaros isoform (Ik6) with dominant negative activity [11,17-19]. Classical techniques allowing detection of microdeletions include whole-genome hybridization-based technologies such as array-CGH and SNP-arrays, and targeted gene-dosage methods such as multiplex-ligation probe assay (MLPA). These techniques estimate the copy-number and therefore have limited sensitivity and they cannot be applied for the detection of minimal residual disease (MRD) in ALL. The deletion breakpoints for these IKZF1Δ4-7 alterations are usually located within a few nucleotides, suggested the feasibility of designing MRD (minimal residual disease) assays based on real-time quantitative polymerase chain reaction (RQ-PCR) in the same way as Ig/TCR-based minimal residual disease (MRD) tests [18]. In this study, in order to detect the levels of IKZF1Δ4-7 alterations, the albumin (ALB) gene was first amplified as the internal control, DNA from the bone marrow (BM) of untreated adult ALL patients from the Chinese mainland were examined by using the TaqMan® probe-based RQ-PCR, multiplex fluorescent PCR and sequence analysis. Simultaneously, the detections of IKZF1Δ4-8, IKZF1Δ2-7 and IKZF1Δ2-8 alterations were also performed.
Patient samples
This study involved 328 newly diagnosed adult ALL patients at Peking University People’s Hospital from 2007 to 2012.At diagnosis and follow up, freshly obtained BM and peripheral blood samples were determined by morphology, flow cytometric immunophenotyping, RQ-PCR BCR/ABL fusion transcript assessment and karyotype analysis. All of the assays were performed by standard routine procedures [20-22]. In addition, IgH-based MRD monitoring was performed and analyzed according to EuroMRD guidelines [23]. Diagnosis was established on the basis of the widely used according as 2008 WHO criteria.
Analysis of IKZF1 deletions
Mononuclear cells were isolated from the BM and peripheral blood samples by using standard Ficoll-Hypaque density gradient centrifugation. DNA was extracted using DNAzol kits (Invitrogen, Carlsbad, CA, USA), according to the manufacturers’ instructions. Deletions in the exons4-7, exons 4-8, exons 2-7, exons 2-8 of the IKZF1 gene (types Δ4-7,Δ4-8,Δ2-7,and Δ2-8) were detected using RQ-PCR. The primers and TaqMan® probe for the albumin (ALB) gene, which was the internal control [24]. The multiplex primers and TaqMan® probes for IKZF1-deletion types Δ4-7, Δ4-8, Δ2-7, and Δ2-8were described as previous reported [18]. Primer sequences for preparation of IKZF1 Δ4-7 plasmid or sequencing IKZF1Δ4-7 deletions were prepared as shown in reference [7]. All the primers and probes used in the study are listed in Table1. The PCR mixture contained 1× TaqMan® Universal PCR Master mix, 400-nM primers, 200-nM fluorescent probes, and 150-500 ng DNA. PCR was performed by using an ABIPRISM® 7500 FAST sequence detection system by employing the following protocol: 50°C for 2 min, 95°C for 10 min, and 45 cycles at 95°C for 15 s and 60°C for 1 min.
Table 1. Primers and probe sequences for detecting IKZF1 deletions
Primer and TaqMan probe sequences for detecting IKZF1 Δ4-7, Δ4-8, Δ2-7 and Δ2-8 deletions |
Δ2a-FP: 5’-CAACAAGTGACCCATCCTTTG-3’
Δ2c-FP: 5’-CTCCTCTAATCTTTGGACTTGTGA-3’
Δ4-FP: 5’-GGAGTCTGTGAAGGTCACACC-3’
Δ7-RP: 5’-AAAGAACCCTCAGGCATTCA-3’
Δ8-RP: 5’-GTCTCGGCATACAGGGAAGA-3’
Δ2a-Probe: 5’-FAM-TTTGCTCAAAAAGGGCACATGTACATTTT-BHQ-3’
Δ2c-Probe: 5’-FAM-AAGGGAGATTGATGTAAGTGGCTCCAC-BHQ-3’
Δ4-Probe: 5’-FAM-TGGTCTTCTCCCAGCCCATAGGG-BHQ-3’ |
Primer and TaqMan probe sequences for detecting ALB control gene |
ALB-FP: 5’-GCC CAT TGT CCT GTT CTG ACT T -3’
ALB-RP: 5’-TTC CAC TGC TGA GCC ATC AC-3’
ALB-Probe: 5’-FAM- TAT GAT GCG GTA CAC AGA GCC ATC CAA G-BHQ-3’ |
Primer sequences for multiplex fluorescent PCR or sequencing IKZF1deletions |
Δ2a-FP: 5’-FAM-CAACAAGTGACCCATCCTTTG-3’
Δ2b-FP: 5’-FAM-CACACACTTCAAGATTATGCATTT-3’
Δ4-FP: 5’-HEX-TGTGAAGGTCACACCCTCTG-3’
Δ7-RP: 5’-AAAGAACCCTCAGGCATTCA-3’
Δ8-RP:5’-GGGGACTGGAAGTCACAGAA-3’ |
Primer sequences for preparation of IKZF1 Δ4-7plasmid standard or sequencing IKZF1Δ4-7 deletions |
C813-FP:5’-CCACAGGGCAAGTCATCCACATTTTG-3’
C814-RP:5’-CAGACCATAGAGTCCCTCCTAGGGGAAAAA-3’
Sequencing C815: 5’-TTCTTAGAAGTCTGGAGTCTGTGAAGGTCA-3’ |
Preparation of plasmid standards
The ALB plasmid standard was obtained as described earlier [24]. IKZF1Δ4-7 was amplified using the DNA isolated from the patients with the IKZF1Δ4-7. The 50-μl PCR mixture contained 25μl of 2× Universal PCR Master mix (TianGen Biotech, Beijing, China), 400 nM primers, and 300 ng DNA. The PCR protocol was as follows: pre-denaturing at 95°C for 5 min, followed by 36 cycles at 95°C for 40 s, 58°C for 40 s, and 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR products were purified and subcloned into the pMD18-T vector (Takara, Dalian, China). Transformation, screening, and sequencing were then performed to obtain IKZF1Δ4-7 plasmid standard. The copy number was calculated based on the optical density (OD) value, and the standards were subjected to a tenfold dilution (107,106,105,104,103,102copies) in order to plot a standard curve. For each measurement, the threshold of amplification for the curve was set at 0.08, and it included a positive control (samples from the patients with the IKZF1Δ4-7), blank control (without template), and negative control (samples from patients without IKZF1Δ4-7).
Calculation of the IKZF1 deletion level
The copy numbers of ALB and IKZF1 deletions were calculated using the Ct value and standard curve. Our study revealed similar efficacies for the amplification of ALB and IKZF1Δ4-7 deletions genes (the slope of standard curve was -3.36±0.15 and -3.46±0.19, respectively). In order to decrease the differences in plasmid quantification, only one ALB standard curve was plotted. The IKZF1 deletions copy number in 100 ALB copies was used as the IKZF1 deletions gene content. If the ALB copy number was ≥3×104, the samples were considered for quantitative detection. The results were analyzed based on the guidelines proposed by the European Study Group for the RQ-PCR detection of MRD [25].
Breakpoint-specific multiplex fluorescent PCR
Multiplex fluorescent PCR was completed as previously described [18]. Primers were designed that they could be combined in a single multiplex PCR and that the amplicons length and fluorescent labelling allowed direct identification of each type of deletion. DNA were amplified in a final volume of 25μl containing 2× Universal PCR Master mix (TianGen Biotech, Beijing, China), 400 nM of each primer, and 150-500 ng DNA. The PCR protocol was as follows: pre-denaturing at 95°C for 5 min, followed by 30 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 10 min. PCR products were run on an ABI 3500 analyser using a fragment size analysis program and analyses were performed using GeneMapper software (Applied Biosystems). The purified PCR products were subjected to direct sequence analysis with an ABI3700 DNA sequencer as described earlier. The screened complicated mutants were subcloned into pMD18-T vectors, followed by transformation, screening and further sequencing.
Statistical analysis
Statistical analysis was performed using SPSS software version 13.0 (Chicago, IL, USA). Independence of categorized parameters was calculated using Chi-square test (or Fisher Exact test). Distribution of continuous variables was calculated using Wilcoxon two sample tests. Values of P<0.05 were considered to be significant.
IKZF1 deletions detection sensitivity
Plasmid standards of ALB and IKZF1Δ4-7 were prepared. Plasmid DNA concentration was determined by absorbance measurement; six serial plasmid dilutions (107,106, 105, 104, 103, and 102 copies) were amplified by RQ-PCR to construct a standard curve for the absolute quantitative assessment of copy number. The results for amplified the ALB and IKZF1Δ4-7 plasmids revealed that all correlation coefficients were above 0.99; the average Ct values for the amplified IKZF1Δ4-7 deletions with copy numbers of 107-101 copies were 15.20±0.14, 18.43±0.08, 21.72±0.32, 25.46±0.16, 28.65±0.24, 32.55±0.33,and 36.04±0.72, respectively (Figure1), and the coefficients of variation were0.9%, 0.5%, 1.5%, 0.6%, 1.6%, 0.8%,1.0%,and 2.0%,respectively; the sensitivity of detection was at least ten copies (Figure 2). In eight DNA samples of IKZF1 Δ4-7-positive patients, the Ct value was in the linear range for a tenfold dilution series (100-10−5) ,the deletions were detected with a sensitivity of 10−4 or 10−5,and the correlation coefficient was above 0.99 (Table 2). High correlation coefficients allowed accurate assessment of the quantity of IKZF1 deletions in unknown samples. All amplification plots were analyzed by positioning the threshold at 0.08.
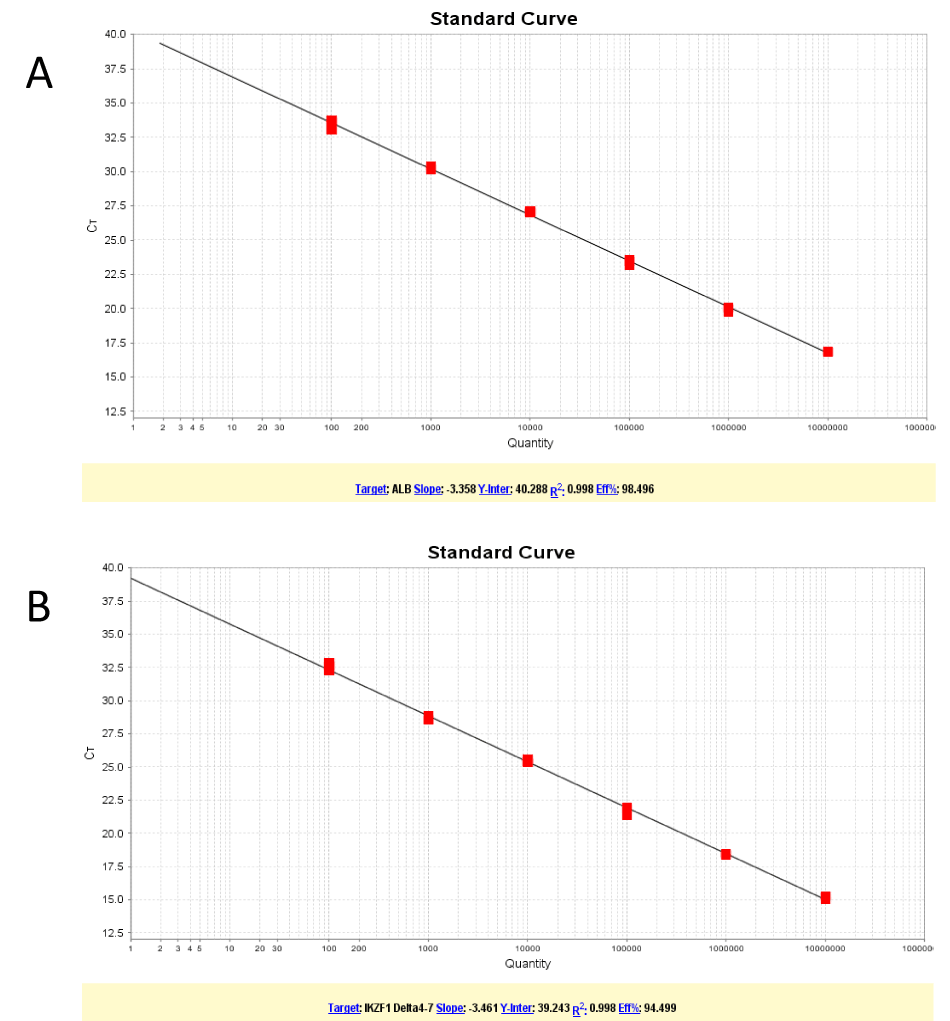
Figure 1. Standard curves obtained by real-time quantitative polymerase chain reaction.(A) ALB gene.(B) IKZF1Δ4-7 deletion
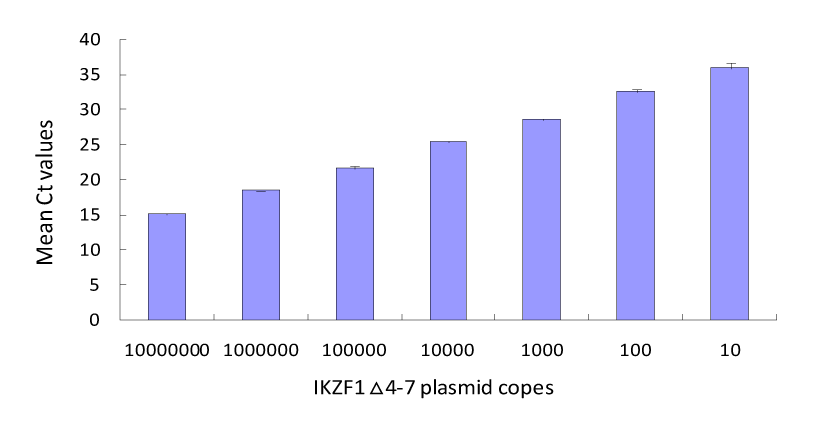
Figure 2. Representative amplification plot of the seven plasmid dilutions of IKZF1Δ4-7 deletion. The sensitivity for detection was at least ten copies. The coefficient of variation (CV) of the Ct value at each dilution was <3%
Table 2. Features of the amplification curves obtained with a tenfold dilution series for patients with IKZF1 deletions
No. of patients |
Deletion |
Maximal |
Mean Ct of |
Mean Ct of maximal |
Dilution |
Correlation |
reproducible |
undiluted |
reproducible |
curve slope |
coefficient |
52 |
△4-7 |
-4 |
23.6 |
37.5 |
-3.6 |
0.999 |
62 |
△4-7 |
-5 |
22.0 |
37.3 |
-3.1 |
0.995 |
31 |
△4-7 |
-4 |
22.3 |
35.9 |
-3.5 |
0.997 |
64 |
△4-7 |
-4 |
23.2 |
37.1 |
-3.4 |
0.998 |
65 |
△4-7 |
-5 |
22.9 |
39.2 |
-3.2 |
0.998 |
79 |
△4-8 |
-4 |
22.3 |
35.6 |
-3.3 |
0.996 |
102 |
△2-7 |
-4 |
22.0 |
35.6 |
-3.4 |
0.999 |
113 |
△2-8 |
-4 |
22.2 |
36.8 |
-3.7 |
0.999 |
Detection of IKZF1 deletions with RQ-PCR
Of the entirety, 36.3% [31.1-41.7%] showed IKZF1-deletion, including 66 (68.8%) with BCR/ABL while 46 (22.4%) without. In 119 IKZF1-deletion positive specimens, the average IKZF1-deletion copies/ALB copy was 138.5% (0.1%-697.9%). The result of distribution for IKZF1-deletion levels in patients with ALL was shown in Table 3. Of 96(80.7%) patients hold higher levels of IKZF1-deletion (12.5%-697.9%),which might be a helpful condition for MRD assay based the IKZF1-deletion.27 IKZF1-deletion-positive patients (219 samples in total) were followed up after treatment, among them, 18 patients in hematologic remission continued to test negative for IKZF1-deletion within 8-66 weeks of follow-up (Figure 3: A). Elevated levels of IKZF1 deletion were detected in five cases when relapse occurred. In four non responders, IKZF1 deletion maintained at high level (Figure 3: B).
Table 3. Distribution for IKZF1-deletion levels in patients with ALL
IKZF1/ALB(%) |
Positive(%) |
0.1-1.0 |
13 (10.9) |
1.1-10.0 |
10 (8.4) |
10.1-100.0 |
41 (34.5) |
101.1-1000.0 |
55 (46.2) |
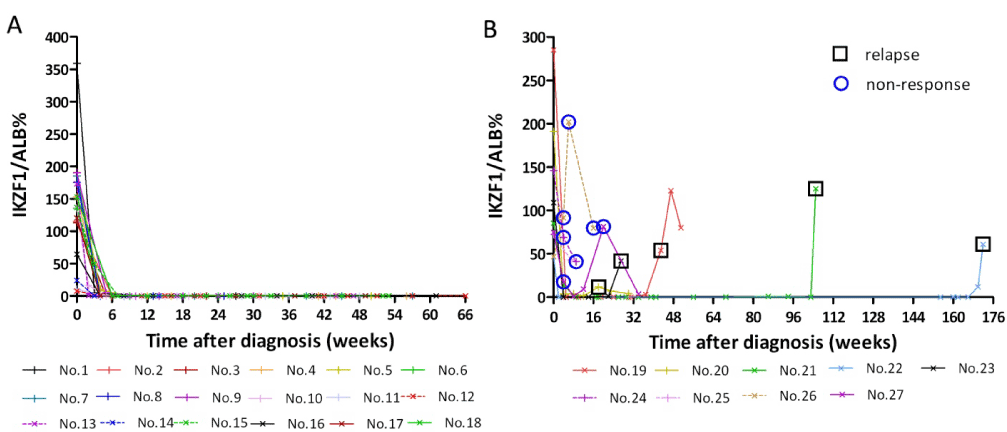
Figure 3. Dynamics of IKZF1 deletions at diagnosis, after induction therapy, and during follow-up in 27 B-ALL patients. (A) No.1-18: Patients in hematologic remission. (B) No.19-23: Patients in relapse; No.24-27: Non responders
MRD analysis was subsequently performed on 48 paired samples from 8 patients and compared with MRD results obtained using IgH markers (Figure 4). The analyses use the log10MRD of the dilution of diagnosis sample giving the same amplification as each sample. A very good correlation (R2 = 0.986, P <0.01) was observed for the quantifiable paired samples. R2 is the Pearson correlation coefficient of the 29 MRD values positive ≥ 10-4 measured by both methods. The results, as previously shown by others for IKZF1 Δ4-7, [17-18] confirm the good accuracy of MRD quantification using IKZF1 markers.
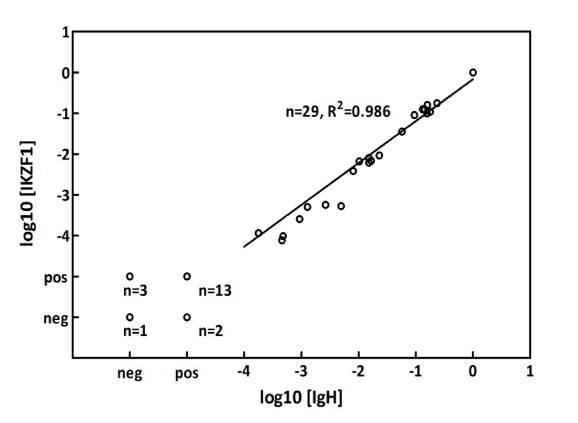
Figure 4. Scatterplot of overall MRD Log10 results using markers based on IKZF1 deletions compared with IgH rearrangements. Detectable but not quantifiable MRD under the 10-4 threshold is termed “pos <10-4” and the opposite “neg <10-4” for undetectable MRD measurement. R2 is the Pearson correlation coefficient of the 29 MRD values positive ≥ 10-4 measured by both methods
Multiplex fluorescent PCR and sequencing results
Multiplex fluorescent PCR and sequence analysis revealed that the 119 positive cases included 80 (67.2%) cases with type Δ4-7, 29 (24.4%) cases with type Δ2-7, seven (5.9%) cases with type Δ4-8, three (2.5%) cases with type Δ2-8, and six cases with deletion of both alleles. In the 119 positive cases, one case with type Δ2-7 and five cases with type Δ4-7 were detected by the multiplex fluorescent PCR and multiplex RQ-PCR but not direct sequencing possibly due to low level of the deletion. The four kinds of IKZF1 deletions with type Δ4-7, Δ2-7, Δ4-8, and Δ2-8 were shown with arrows indicating the region in which the breakpoints occur (Figure 5). Genomic breakpoint sequences of the IKZF1 deletion in the 113 ALL cases were described in the supplementary appendix.
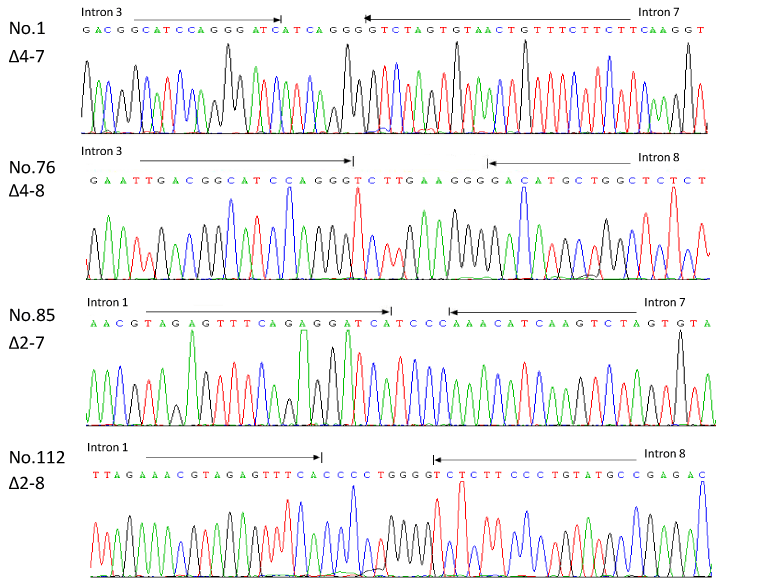
Figure 5. Genomic breakpoint sequences of the IKZF1 deletions in the four ALL cases. Four kinds of IKZF1 deletions are shown with arrows indicating the region in which the breakpoints occur. Regions matching the reference genomic IKZF1 sequence are shown by arrows, separated by additional nucleotides not matching the consensus sequence
Clinical Characteristics of IKZF1 deletions-positive patients
The characteristics of IKZF1-deleted vs. wild-type patients are shown in Table 4. Patients with B-ALL had a higher IKZF1-deletion rate (118/292, 40.4%) than those with T-ALL (1/36, 2.8%,P<0.01).Common B-cell ALL group had higher IKZF1 deletions rate(96/196,49.0%) than those with progenitor B-cell ALL(12/41,29.3%,P<0.05) or precursor B-cell ALL (10/55,18.2%,P<0.01).Cytogenetic data were available in 290 patients. The detection rates of IKZF1 deletions-positive mutations in patients harboring Ph chromosome/ (BCR/ABL)-transcripts group and non harboring group were 68.8% (66/96) and 22.4% (46/205), respectively, showing a significant difference (P<0.01). Patients with other-karyotypes-present which involved the cases with complex karyotype, MLL-translocation and near triploidy or del (9p) except of t(9;22)/(BCR/ABL)did not show a higher IKZF1 deletion rates. Moreover, IKZF1 deletions were significantly associated with age above 35, higher initial white blood cell counts (P<0.01),but not with gender, blast in the bone marrow, hemoglobin and platelet counts in the peripheral blood.
Table 4. IKZF1-deletion in relation to clinical and laboratory features in adult ALL patients
|
Variant
|
Total(n=328)
|
IKZF1-deleted(n=119, 36.3%)
|
Wild type(n=209, 63.7%)
|
P
|
|
n (%)
|
n(%)
|
n(%)
|
|
Age, years
|
|
Median(range)
|
32(14-80)
|
37(14-72)
|
28(14-80)
|
0.020
|
|
low <35
|
180(54.9)
|
52(28.9)
|
128(71.1)
|
P<0.01(low vs. high)
|
|
high ≥35
|
148(45.1)
|
67(45.3)
|
81(54.7)
|
|
Gender
|
|
|
|
|
|
Male
|
179(54.6)
|
57(31.8)
|
122(68.2)
|
P>0.05
|
|
Femal
|
149(45.4)
|
62(41.6)
|
87(58.4)
|
|
|
BM Blast(%)
|
|
75.2
|
81.2
|
0.458
|
|
WBC count,×109/L
|
75.2
|
44.9
|
0.006
|
|
Hemoglobin(g/L)
|
86.6
|
95.1
|
0.160
|
|
Platelet,×109/L
|
75.1
|
75.1
|
0.997
|
|
t(9;22)/BCR-ABL1
|
|
|
|
|
Present
|
96(29.3)
|
66(68.8)
|
30(31.3)
|
P<0.01
|
|
Absent
|
205(62.5)
|
46(22.4)
|
159(77.6)
|
|
|
NAa
|
27(8.2)
|
7(25.9)
|
20(74.1)
|
|
|
Others karyotypesb
|
|
Present
|
124(37.8)
|
33(26.6)
|
91(73.4)
|
P<0.01
|
|
Absent
|
166(50.6)
|
74(44.6)
|
92(55.4)
|
|
|
NAa
|
38(11.6)
|
12(31.6)
|
26(68.4)
|
|
|
Immunologic subtype
|
|
B-ALL
|
292(89.0)
|
118(40.4)
|
174(59.6)
|
P<0.01c
|
|
Pro-B-ALL
|
41(14)
|
12(29.3)
|
29(70.7)
|
P<0.05d
|
|
Pre-B-ALL
|
55(19)
|
10(18.2)
|
45(81.8)
|
P<0.01d
|
|
Com-B-ALL
|
196(67)
|
96(49.0)
|
100(51.0)
|
|
|
T-ALL
|
36(11.0)
|
1(2.8)
|
35(97.2)
|
|
2021 Copyright OAT. All rights reserv
a Not available/not evaluated due to <20 metaphases.
b Including cases with complex karyotype, MLL-translocation, near triploidy, or del(9p) but except t(9;22)/(BCR/ABL).
c Compared to T-cell ALL group.
d Compared to Common B-cell ALL group.
RQ-PCR shows the IKZF1-deletion rate was as high as 36.3% in the untreated 328 ALL patients and the combination rate of BCR/ABL was more frequent than not(68.8% vs.22.4%, P<0.01), as previously published data shown [11,15,26-28]. Patients with B-ALL had a higher IKZF1-deletion rate than those with T-ALL, especially, our result showed that IKZF1-deletion rate in common B-cell ALL was the highest among all B-ALL subtype. Sequence analysis and multiplex fluorescent PCR revealed that 125 IKZF1 intragenic deletions were detected in 119 patients (6 patients having deletion of both alleles), all of which were Δ4-7, Δ2-7, Δ4-8 or Δ2-8. The detection rate of type Δ4-7 in our study was the highest among the four kinds of intragenic deletions, as previously shown by others [17-19, 27-28].
MRD analysis was subsequently performed on 54 paired samples and compared with MRD results obtained using IgH markers (Figure 2). A very good correlation (R2 = 0.986, P<0.01) was observed for the quantifiable paired samples. The results, as previously shown by others for IKZF1 Δ4-7 [18-19], confirm the good accuracy of MRD quantification using IKZF1 markers. They examined reproducibility measurement of MRD using markers based on IKZF1 deletions compared with Ig/TCR rearrangements and found that the IKZF1 results were as close to the Ig/TCR results. It is therefore likely that this IKZF1 marker will be at least as stable as Ig/TCR rearrangements, although this will need to be confirmed in more extensive studies. Two DNA MRD markers with high sensitivity (at least 10-4) are generally required in MRD intervention clinical trials [29-30], and in a large cohort of 2854 pediatric precursor B-cell ALL patients, 20% of patients had only one sensitive marker and 8% had none [30]. At present the concept of using disease-related markers for MRD testing has been already established for fusion transcripts such as BCR-ABL and for gene rearrangements such as for SIL-TAL1 in T-ALL and for MLL rearrangements in infant ALLs [31]. The use of oncogenic lesions for MRD monitoring has been limited by the fact that recurrent chromosomal translocations are not found in all ALL and are usually studied at the RNA level, so that IKZF1 gene deletions will provide a useful addition to the repertoire of MRD markers currently available for monitoring MRD in ALL and inclusion of this marker in standard screening for MRD targets would be an easy way to provide more patients with two sensitive markers.
By using ALB as the internal control, we found that in 119 IKZF1-deletion positive patients, the average IKZF1-deletion copies/ALB copies was 138.5% (0.1%-697.9%) at diagnosis. Of 96(80.7%) patients hold higher levels of IKZF1-deletion (12.5%-697.9%), which might be a helpful condition for MRD assay based the IKZF1-deletion.27 IKZF1-deletion-positive patients (219 samples in total) were followed up after treatment, among them, 18 patients in complete hematologic remission maintained in negativity for IKZF1-deletion within 8-66 weeks. In five relapsed cases, elevated levels of IKZF1 deletion were detected when relapse occurred. In the four non responders, IKZF1 deletion maintained at high level. Kuiper et al. [32] in an analysis of paired diagnosis and relapse samples from 34 patients found IKZF1 deletions at diagnosis and showed that all were conserved at relapse, in contrast to other recurrent genetic lesions found at diagnosis such as PAX5, CDKN2A and EBF1. Therefore, to detect the levels of IKZF1-deletion during the clinical course would be helpful for monitoring early relapse cases in ALL patients.
In summary, we found that RQ-PCR based on ALB as control gene was a reliable and sensitive method for detecting IKZF1 deletions. Consensus breakpoint sequences can be used as markers for MRD monitoring. This work should be useful for adjunctive diagnosis, risk assessment and studying the underlying pathogenesis of adult B-ALL, especially in common-B-cell ALL.
The authors report no declarations of interest.
The study design adhered to the principles of the Helsinki Declaration and was approved by the ethics committees of our hospital.
Informed consent was obtained from all the patients prior to their enrollment in the study.
GRR designed the project and prepared the typescript. LXW,JZ and QMY performed the experiments and the analyses. JLL, LDL, CY, NL, XL, YZQ, YRL, YYL, SSC, KYL and XJH fulfilled ethical authorizations, collected and stored data. All authors approved the typescript.
This study was supported by the National Basic Research Program of China#1 under Grant number 2013CB733701, the Key Program of National Natural Science Foundation of China #2 under Grant number 81230013, the National Natural Science Foundation of China#3 under Grant number 81570182 and the Specialized Research Fund for the Doctoral Program of Higher Education of China#4 under Grant number 20130001110079.
View supplementary data
- Gleissner B, Thiel E (2003) Molecular genetic events in adult acute lymphoblastic leukemia. Expert Rev Mol Diagn 3: 339-355.
- Armstrong SA, Look AT (2005) Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol 23: 6306-6315. [Crossref]
- Pui CH, Relling MV, Downing JR (2004) Acute lymphoblastic leukemia. N Engl J Med 350: 1535-1548. [Crossref]
- Moorman AV, Harrison CJ, Buck GA, Richards SM, Secker-Walker LM, et al. (2007) Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): analysis of cytogenetic data from patients treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. Blood 109: 3189-3197. [Crossref]
- Bungaro S, Dell’Orto MC, Zangrando A, Basso D, Gorletta T, et al. (2009) Integration of genomic and gene expression data of childhood ALL without known aberrations identifies subgroups with specific genetic hallmarks. Genes Chromosomes Cancer. 48: 22-38.
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, et al. (2007) Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446: 758-764. [Crossref]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, et al. (2008) BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453: 110-114. [Crossref]
- Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, et al. (1994) The Ikaros gene is required for the development of all lymphoid lineages. Cell 79: 143-156. [Crossref]
- Kuiper RP, Waanders E, van der Velden VH, van Reijmersdal SV,Venkatachalam R, et al. (2010) IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia 24: 1258-1264. [Crossref]
- Moorman AV (2012) The clinical relevance of chromosomal and genomic abnormalities in B cell precursor acute lymphoblastic leukaemia. Blood Rev 26: 123-135. [Crossref]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, et al. (2009) Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 360: 470-480. [Crossref]
- Giovanni Martinelli, Ilaria Iacobucci, Clelia Tiziana Storlazzi, Marco Vignetti, Francesca Paoloni, et al. (2009) IKZF1 (Ikaros) Deletions in BCR-ABL1-Positive Acute Lymphoblastic Leukemia Are Associated With Short Disease-Free Survival and High Rate of Cumulative Incidence of Relapse: A GIMEMA AL WP Report. Journal of Clinical Oncology 27: 5202-5207.
- Waanders E, van der Velden VH, van der Schoot CE, van Leeuwen FN, vanReijmersdal SV, et al. (2011) Integrated use of minimal residual diseaseclassification and IKZF1 alteration status accurately predicts 79% of relapses inpediatric acute lymphoblastic leukemia. Leukemia 25: 254-258. [Crossref]
- Harvey RC, Mullighan CG, Chen IM, Wharton W, Mikhail FM, et al. (2010) Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood 115:5312-5321. [Crossref]
- Chen IM, Harvey RC, Mullighan CG, Gastier-Foster J, Wharton W, et al. (2012) Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children's Oncology Group study. Blood 119: 3512-3522. [Crossref]
- Yao QM, Liu KY, Gale RP, Jiang B, Liu YR, et al. (2016) Prognostic impact of IKZF1 deletion in adults with common B-cell acute lymphoblastic leukemia. BMC Cancer 16: 269. [Crossref]
- Meyer C, Zur Stadt U, Escherich G, Hofmann J, Binato R, et al. (2013) Refinement of IKZF1 recombination hotspots in pediatric BCP-ALL patients. Am J Blood Res 3: 165-173. [Crossref]
- Aurélie Caye, Kheïra Beldjord, Kelly Mass Malo, Séverine Drunat, Jean Soulier, et al. (2013) Breakpoint-specific multiplex PCR allows the detection of IKZF1 intragenic deletions and minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia. Haematologica 98: 597-601. [Crossref]
- Venn NC, van der Velden VH, de Bie M, Waanders E, Giles JE, et al. (2012) Highly sensitive MRD tests for ALL based on the IKZF1 Δ3-6 microdeletion. Leukemia 26: 1414-1416. [Crossref]
- Liu YR, Yu H, Chang Y, Chen SS. (2003) The application of 4-color fluorescence labeling antibodies and its significance in immunophenotyping for leukemia by flow cytometer. Zhongguo Shi Yan Xue Ye Xue Za Zhi 10: 423-427. [Crossref]
- Qin Y-Z, Liu Y-R, Li J-L, Fu J-Y, Chang Y, et al. (2003) Follow-up Detection of M-bcr/abl and m-bcr/ abl fusion transcripts in chronic myeloid leukemia. Ann Hematol patients after allogeneic hematopoietic stem cell transplantation. Zhongguo Shi Yan Xue Ye Xue Za Zhi 11: 368-371.
- Zhang Y, He Q, Huang XJ, Jiang H, Yang SM, et al. (2007) [Cytogenetic study on eosinophilia]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 15: 454-457. [Crossref]
- van der Velden VH, Cazzaniga G, Schrauder A, Hancock J, Bader P, et al. (2007) Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative data. Leukemia 4: 604-611. [Crossref]
- Ruan GR, Li JL, Qin YZ, Li LD, Xie M, et al. (2009) Nucleophosmin Mutations in Chinese Adults with Acute Myelogenous Leukemia. Annals of Hematology 88:159-166.
- Van der Velden VH, Hochhaus A, Cazzaniga G, Szczepanski T,Gabert J, et al (2003) Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: principles, approaches, and laboratory aspects. Leukemia 17: 1013-1034. [Crossref]
- Iacobucci I, Storlazzi CT, Cilloni D, Lonetti A, Ottaviani E, et al. (2009) Identification and molecular characterization of recurrent genomic deletions on 7p12 inthe IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemiapatients: on behalf of Gruppo Italiano Malattie Ematologiche dell'Adulto AcuteLeukemia Working Party (GIMEMA AL WP). Blood 114:2159-2167. [Crossref]
- Iacobucci I, Iraci N, Messina M, Lonetti A, Chiaretti S, et al. (2012) IKAROS deletions dictate a unique gene expression signature in patients with adult B-cell acute lymphoblastic leukemia. PLoS One 7: e40934. [Crossref]
- Tokunaga K, Yamaguchi S, Iwanaga E, Nanri T, Shimomura T, et al. (2013) High frequency of IKZF1 genetic alterations in adult patients with B-cell acute lymphoblastic leukemia. European journal of haematology 91: 201-208.
- Brüggemann M, Schrauder A, Raff T, Pfeifer H, Dworzak M, et al. (2010) Standardized MRD quantification in European ALL trials: proceedings of the Second International Symposium on MRD assessment in Kiel, Germany, 18-20 September 2008. Leukemia 24: 521-535. [Crossref]
- Flohr T, Schrauder A, Cazzaniga G, Panzer-Gru¨mayer R, van der Velden V, et al. (2008) Minimal residual disease (MRD)-directed risk stratification using real-time quantitative PCR analysis of immunoglobulin and T-cell receptor gene rearrangements in the international multicenter trial AIEOP-BFM ALL 2000 for childhood acute lymphoblastic leukemia (ALL). Leukemia 22: 771-782. [Crossref]
- van der Velden VHJ, Corral L, Valsecchi MG, Jansen MWJC, De Lorenzo P, et al. (2009) Prognostic significance of minimal residual disease in infants with acute lymphoblastic leukemia treated within the Interfant-99 protocol. Leukemia 23: 1073-1079. [Crossref]
- Kuiper RP, Waanders E, van der Velden VH, van Reijmersdal SV, Venkatachalam R, et al. (2010) IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia 24: 1258-1264. [Crossref]





