Abstract
G protein-coupled receptors (GPCRs) constitute the largest protein superfamily in mammalian genomes. The Leucine-rich repeat-containing G protein-coupled receptors LGR4, LGR5 and LGR6 belong to the type A rhodopsin-like family and are closely structurally related to the glycoprotein hormone receptors. They have the particularity to be expressed in stem/progenitor cells. LGRs commonly recognize R-spondins as ligands leading to Wnt signaling regulation. Whereas LGR4 plays an essential role during development and adult homeostasis and exerts a dominant function over its paralogues, the function of LGR5 still remains controversial. In cancer cells, LGRs identify tumor-initiating cells and their expression can be correlated with tumor stage and prognosis. The molecular events underlying deregulated expression of LGRs in cancer cells and potential new therapeutic approaches to target cancer cells are reviewed.
Key words
LGR4, LGR5, LGR6, stem cells, cancer, R-spondins, GPCR, Wnt
Introduction
The Leucine-rich repeat-containing G protein-coupled receptors LGR4/GPR48, LGR5/GPR49 and LGR6 were identified in the late nineties as new members of the large G protein-coupled receptor (GPCR) superfamily within the subfamily of LGR receptors, which exhibit large ectodomains composed of a variable number of Leucine-rich repeats (LRR) (Figure 1A). The LGR4/LGR5/LGR6 receptors (LGR subfamily group B) contain a large extracellular region (17 LRRs) as compared to the one of glycoprotein hormone receptors (TSHR/FSHR/LHR-group A) or Relaxin/Insulin-like receptors (LGR7/LGR8-group C) only made of 9 LRRS [1-3]. LGR4 shares 46 and 44% overall similarity with LGR5 and LGR6, respectively whereas LGR5 and LGR6 are more closely related together (54% similarity). From an evolutionary point of view, the three human LGR4/LGR5/LGR6 paralogues, highly conserved in mammalians (90%, 82% and 84% similarity with the mouse orthologues Lgr4/Lgr5/Lgr6, respectively), are also found in teleosts as two paralogues (LGR4 and LGR6) and in invertebrates as one single putative ortholog DLGR2 [4]. Primary structure analysis and crystallography studies suggest that LGR4/LGR5/LGR6 share a conserved LRR-NT domain protecting the LRR1 from solvent exposure [5]. Of interest, their LRR-CT region (also designated Hinge region), lying between the LRRs and the seven transmembrane (7TM) domain, contain the YXXXCC and the FXPCE motifs, modules highly conserved within the glycoprotein hormone receptors and involved in receptor activation [3,6]. Similarly, the LXFT or NPXXY motifs in the transmembrane domains TM6 and TM7, recognized as important for GPCR activity, are also conserved in LGR4/LGR5/LGR6, indicating that these receptors exhibit several characteristics of classical GPCRs, and thus might function as such [3] (Figure 1A). The LGRs were deorphanized in 2011. It was demonstrated that the three LGRs can redundantly recognize the 4 members of the Rspondin family as ligands and that this interaction strongly potentiates the Wnt/β-catenin canonical pathway in vitro [7-9] (Figure 1A). Unexpectedly, ligand binding does not trigger canonical G protein-dependent activation of these receptors [7-10]. Thus, LGRs appear as peculiar GPCRs playing a pivotal role in the regulation of Wnt signaling. The scope of this review is to summarize the main findings obtained so far about the role of LGR receptors in vivo and about their potential relation to cancer.
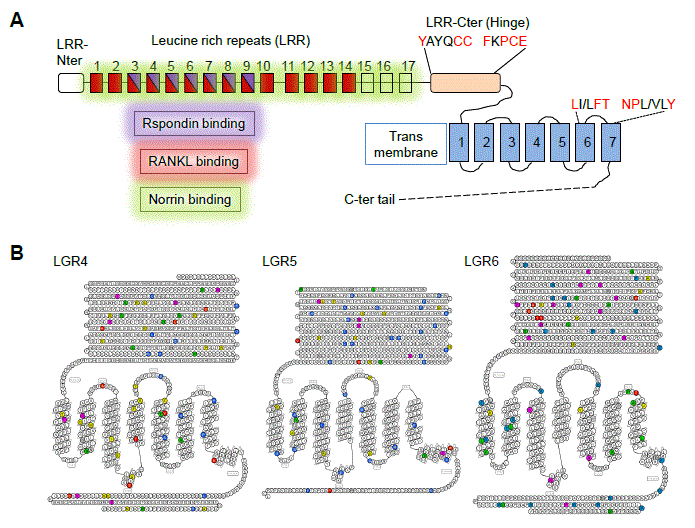
Figure 1. schematic representation of LGR4/5/6 receptors. A. Structure and function of the LGR receptors. The Leucine-rich repeats (LRR) are represented as rectangles and areas of Rspondins, RANKL and Norrin binding are highlighted in purple, red and green, respectively. GPCR consensus residues present in the hinge and transmembrane domains are evidenced in red. B. Snake diagram of the LGR4, LGR5 and LGR6 receptors. Residues associated with somatic mutations are colored : colorectal cancer (yellow), stomach cancer (green), melanoma (blue), uterine cancer (pink), breast cancer (red) and head and neck cancer (grey).
Lgrs and in vivo functions
LGR4
Tissue expression and in vivo function: LGR4 is the member of the family whose pattern of expression and function have been the most extensively studied. This receptor is widely and abundantly expressed in mouse tissues originating from the three embryonic layers, in the regions known to contain progenitors and stem cells [11]. In vivo studies have demonstrated that this receptor plays an essential role during development as Lgr4-deficient embryos display embryonic and perinatal lethality [12]. Moreover, developmental defects have been reported for homozygous mice in many organs, including among others, altered tubulogenesis, impaired branching morphogenesis, renal hypoplasia, corneal dysgenesis or gallbladder agenesis [13-25]. In line with expression of Lgr4 in progenitors/stem cells, its deficiency generally correlates with reduced cell proliferation in tissues [7,15,19,23,26-29]. Moreover, though premature differentiation has been reported in Lgr4-deficient embryos [17,21,30], Lgr4 deficiency has been mainly described to be associated with impaired or retarded cell differentiation [23,24,28,29,31-33]. Such phenotypes have been demonstrated to be associated with decreased Wnt signaling in intestinal, liver and dental epithelia as well as in peritubular myoid cells in testis [7,23,28,31,34,35]. Accordingly, experiments using GSK3b inhibitors (LiCl, CHIR99201) or in an Apcmin/+ background efficiently restored Wnt activity in the intestine of Lgr4-knockout homozygous mice ex vivo and in vivo [7,28,34].
Molecular mechanisms associated with LGR4 function: Several reports have investigated the means by which this receptor regulates the Wnt pathway, principally in vitro. Wnt signaling must be fine-tuned in stem cells to control the balance between self-renewal and differentiation. The current model is that Wnt ligand binding to the coreceptor complex Frizzled-LRP5/6 at the cell membrane induces LRP5/6 phosphorylation and signalosome formation by local recruitment of the scaffold protein Dishevelled (Dvl) that inactivates the b-catenin destruction complex consisting of glycogen synthase kinase 3, casein kinase I, adenomatosis polyposis coli, axin and an E3 ubiquitin ligase b-Trcp. This process results in stabilization and accumulation of intracellular b-catenin pools, which then reach the nucleus to activate transcription of Wnt/b-catenin target genes [36]. In turn, among these Wnt target genes, the related membrane-bound E3 ubiquitin ligases Znrf3 and Rfn43 play an essential role to suppress Wnt/b-catenin signaling. Binding of Znrf3/Rfn43 to the complex Frizzled-LRP5/6 induces Wnt receptor degradation via ubiquitination leading to attenuation of Wnt signaling [37,38]. Since Rspondins bind to Znrf3/Rfn43 with low affinity, it is suggested that the interaction of Rspondin with LGR4 and Znrf3/Rfn43 enhances the clearance of the ubiquitin ligase from the cell membrane to potentiate Wnt signaling [37,39]. Moreover, in presence of Rspondin, LGR4 is stimulated to interact via its 7TM domain with the IQGAP1/2/3 signaling molecules, increasing the affinity of IQGAPs for the scaffold protein Dvl in the Wnt signalosome complex [39]. Accordingly, LGR4 has been detected in supercomplexes containing the Fzd and Lrp5/6 receptors [7]. Overall, the Rspondin/LGR4/IQGAP1 axis enhances Lrp5/6 phosphorylation through local engagement of a MEK kinase, leading to intracellular b-catenin stabilization and thus potentiation of Wnt/b-catenin signaling [39]. In addition, in line with in vivo studies showing that Lgr4 mediates PCP signaling in Xenopus, interaction of Rspondin-LGR4 with the pivotal protein IQGAP1 can also activate the non-canonical Wnt signalosome by enhancing focal adhesion assembly and cell migration [9,39]. Beside the Rspondin ligands, the Norrin protein belonging to the bone morphogenic protein antagonist family has also been described to bind the three LGR paralogues, but only interaction with LGR4 does stimulate the Wnt/b-catenin pathway (nevertheless independently of Wnt3a or Rspondins) [40] (Figure 1A). In contrast, interaction of LGR4 with the RANKL ligand (also known as Tumor necrosis factor superfamily member 11-Tnfsf11) involves intracellular Gaq engagement leading to inhibition of RANKL-induced osteoclast activation and bone loss [41]. This suggests that LGR4 would act as a decoy receptor for RANKL, a function which can be suppressed by Rspondin1/Rspondin2 due to competitive partially overlapping binding of these two ligands to the LGR4 ECD (Figure 1A). Still, controversy remains regarding potential involvement of the Wnt/b-catenin in this process [41,42]. Another argument for LGR4 activity regardless of Wnt/b-catenin stimulation is the reported control of LGR4 in the white-to-brown fat switch in adipose tissues. This involves LGR4 activation and downstream cAMP/PKA/CREB signaling involving a conserved GPCR 7TM residue (A750) [43]. Of note, the gain-of-function A750T (c.2248 G>A) variant has been associated with human central obesity [44]. Similarly, an LGR4-cAMP/PKA axis activation cascade has been reported by the same group during development in the eye, fetal liver and adrenal gland [13,27,32]. Although still an open question, the LGR4 receptor may exert its function on stem/progenitor cells by means of different cascades depending on the cell type.
LGR5
Tissue expression and in vivo function: LGR5 is described as a Wnt/b-catenin target gene [45]. Accordingly, its expression is essentially restricted to adult stem cells in high rate self-renewing tissues like intestine, stomach, colon and skin [45-47]. In other tissues such as kidney, its expression, detected during tissue morphogenesis becomes silenced in adulthood meanwhile LGR5 expression is induced in injured pancreas via a Wnt-dependent regenerative process in adults [48,49]. In the mammary gland, Lgr5-expressing cells are sufficient and necessary for gland organogenesis [50]. The Lgr5 progeny population switches from the luminal to the myoepithelial compartments during postnatal development of this gland [51]. In digits, Lgr5 expression is detected in the dermis [52]. Of relevance, Lgr5 is detected in cells also expressing the paralogue receptors Lgr4 and/or Lgr6, thus rendering the analysis of individual receptor function quite complex [7,28,30,35,53]. In vivo function studies in adults have been hampered by the neonatal lethality of plain null embryos that present an ankyloglossia phenotype [54]. Nevertheless, in the intestine, Lgr5 null embryos exhibit precocious Paneth cell differentiation associated with increased crypt stem cell- and Wnt target gene markers, in marked contrast with the low Wnt signaling tone observed in Lgr4 null embryos/mice [28,29,55]. Similarly, in the embryonic cochlea, Lgr5 deficiency leads to hair cell overproduction associated with Wnt/b-catenin activity [30]. Consistant with non-redundant activity of these two receptors in some tissues, rescue of the Lgr5 null-ankyloglossia phenotype was observed in double knockout Lgr4/Lgr5 mice, allowing their neonatal survival [28]. Conversely, in skin and kidney where Lgr5 null embryos show a subtle phenotype as compared to the dominant cognate Lgr4 receptor, double knockout Lgr4/Lgr5 embryos exhibit a worsened phenotype, suggesting that these two receptors both contribute to the stem cell pools in these tissues [28,29]. Similarly, in adult intestine and liver, conditional ablation of Lgr5 has no overt phenotype but conditional ablation of both Lgr4 and Lgr5 receptors further aggravates the phenotype induced by Lgr4 deficiency [7,35]. In sum, in the absence of additional data on the spectrum of the regulatory cascades they control, the question of functional redundancy of Lgr4 and Lgr5 in vivo is still open.
Molecular mechanisms associated with LGR5 function: As compared to its paralogues that are not direct Wnt target genes, Lgr5 expression is induced by Wnt signaling in a suggested bi-modal manner [56-58]. Expression of the receptor would occur under a narrow range of “not too low, not too high” Wnt stimulation, which may explain in part why the Lgr5 function is still subject to discussion. The interaction of Lgr5 with Rspondin ligands has been extensively studied by crystallography on Rspondin/LGR4/LGR5 ectodomain complexes [5,59,60]. Rspondins are multifunctional proteins made of several domains. The two Furin domains, organized as Cystein-knotted b hairpin structures, are the ones involved in LGR binding [7,9]. The overall contact surface area is 800 Å2 and involves 2 binding pockets (one hydrophilic and one hydrophobic) lying within the extracellular domains LRR3 to LRR9. These regions are shared by the 3 LGRs and specific residues have been demonstrated to be critical for Wnt activation on cell lines [5]. Analysis of the ternary complex LGR5/Rspondin/Rnf43 cristals shows that LGR5 does not directly interact with the E3 ubiquitin ligase Rnf43 [5,60]. On HEK293T cells overexpressing the LGR5 receptor, evidences have been provided that Rspondin activates Lgr5 to potentiate Wnt/b-catenin activity by enhancing its interaction with the Lrp6 and Fzd5 co-receptors [61]. In turn, such association with LGR5 then accelerates the internalization, endocytosis and degradation of these receptors resulting in Wnt desensitization [8,61]. In corneal endothelial cells expressing LGR5, such acceleration of b-catenin turnover has also been reported upon Rspondin stimulation [62]. In accordance with these observations, in colorectal cancer cells expressing high levels of LGR5, Rspondin 2 stimulation via this receptor negatively regulates the Wnt/b-catenin pathway and suppresses tumor cell growth, suggesting that under these conditions, the LGR5 receptor would function as a negative regulator of the Wnt pathway [56,63]. Interestingly, such negative feedback loop activity on this pathway has been reported for another Wnt target gene, the Troy (Tumor necrosis Factor receptor family member 19) receptor, which can interact with LGR5 at the cell membrane [64]. Of relevance, LGR5 is constitutively internalized, a process that depends on clathrin and is further stimulated in presence of Rspondin and Wnt ligands [61,65]. Evidences have been provided that the Lgr5 C-terminus region contains phosphorylation sites determining receptor internalization and intracellular trafficking and impacting on Wnt/bcatenin activity[61,65,66]. Altogether, these data indicate that cell surface expression of the LGR5 receptor represents another important parameter regulating LGR5 activity on Wnt signaling. Counter-intuitively, whereas C-terminus deleted-Lgr5 receptors are stabilized at the cell membrane in vitro enhancing the Wnt/bcatenin pathway, their expression in intestinal stem/progenitors is associated with diminished cell fitness as compared to the intact Lgr5 receptor in vivo [67]. These data demonstrate a biological role of the Lgr5 intracellular domain on stemness. Another study suggests that the Hinge region of the LGR5 receptor, located between the LRR repeats and the 7TM, also regulates Wnt/b-catenin activity as antibodies directed against this region strongly enhance Topflash activity even in the absence of Rspondin [60] (Figure 1A). Of interest, a similar Hinge region in the cognate glycoprotein hormone receptors participates to an auto-inhibitory activity on GPCR signaling, meanwhile antibodies targeting this domain induce a conformational change alleviating this inhibitory state [68]. Together, these observations may be compatible with a role of LGR5 regulating Wnt activity by means others than the sole Rspondin interaction via LRRs. Furthermore, additional reports indicate that Lgr5 can activate other cascades. For example, LGR5 has been shown to predominantly signal on a noncanonical Wnt pathway in adrenal gland progenitors leading to inhibition of aldosterone production [69]. In HEK293T cells, Lgr5 expression, but not that of Lgr4/Lgr6, can stimulate the G12/13-Rho GTPase and NFkB pathways in absence of the Rspondin ligand [70]. Altogether, these studies evidence a complex and tightly regulated function of the Lgr5 receptor on stem/progenitors, involving both the extracellular and the intracellular domains of the molecule. The exact contribution of each of these mechanisms to Lgr5 function remains to be investigated.
LGR6
Tissue expression and in vivo function: The third member of the Lgr family, LGR6, is the less characterized in terms of gene expression and function. Dynamic expression of Lgr6 has been reported during cochlear development. First detected in prosensory cells of the middle and basal turn, Lgr6 expression becomes progressively restricted to inner pillar and inner border cells before complete disappearance in adult cochlea [71]. In this tissue, stimulation of Wnt/bcatenin activity can induce re-appearance of Lgr6-expressing cells but it does not directly activate expression of the Lgr6 receptor itself [71]. In the mammary gland, Lgr6 labels rare populations of basal and luminal cells, which behave as unipotent progenitors clonally expanding during puberty and regaining proliferative capacity and generating alveoli during pregnancy [72]. In adults, Lgr6 marks cells with long-term self-renewing capacity in the hair follicle, sebaceous and interfollicular epidermis compartments in the skin [73-77]. In the lung, it is expressed by a discrete population of stem/progenitor cells co-expressing the integrin a6 and the paralogue Lgr5 [78]. In taste buds, Lgr6 marks stem/progenitor cells in both the anterior and posterior tongue whereas Lgr5-expressing cells are only detected in the posterior part [53]. In digit tips, Lgr6 expression is detected in nail stem cells and bone, in a pattern correlating also with Lgr4 expression [52]. Regarding Lgr6 function in vivo, Lgr6 knockout mice are fertile and absence of the receptor in the skin does not impact on cell proliferation, differentiation in sebaceous glands or even on wound healing and cell migration [76]. In contrast, although nails develop normally in absence of Lgr6, they fail to regenerate in some Lgr6-deficient mice after amputation, indicating a contribution of this receptor during digit tip regeneration [52].
Molecular mechanisms associated with LGR6 function: Similar to the cognate Lgr4/Lgr5 receptors, in transfected cells ex vivo, Lgr6 binds all members of the Rspondin family with high affinity and potentiates Wnt/b-catenin activity, in a heterotrimeric G protein classes or b-arrestin-independent manner [7,79]. Nevertheless, the potentiating effect of Lgr6/Rspondin interaction is weaker as compared to the paralogues [79]. Two naturally occurring somatic mutants identified in colorectal cancer (G725C and P928H) were tested in Topflash assays to address their potential impact on Wnt signaling. They turned out to stimulate this pathway at levels similar to the wild-type receptor [79]. In contrast, another cancer-associated insGRS insertion led to loss of Rspondin binding and concomitant reduced Wnt potentiation [79]. This showed that somatic mutations in LGR6 can affect Wnt activity but the possibility that other pathways are regulated by LGR6 still remains an open question.
Lgrs and cancer
The LGR receptors mark stem and/or progenitors in adult tissues and can modulate their self-renewal and/or differentiation ability. As cancer-initiating cells exhibit many characteristics of stem cells, efforts have been made in the last decade to address whether these receptors may also be considered as markers of cancer cells and to investigate the hypothetical function of LGRs in cancer initiation and progression, using in particular mouse model studies. In a colorectal cancer model using a conditional Apc loss-of-function mouse line, specific ablation of Apc function in Lgr5-expressing (Lgr5+ve) stem cells results in cell transformation leading to adenoma development along the small and large intestine [80]. A similar conclusion has been reached with the Apcmin/+ line used as a model for human familial adenomatous polyposis [81]. In addition, overexpression of the Rspondin 3 ligand in the Lgr4+ve/ Lgr5+ve cells also leads to rapid adenoma and adenocarcinoma development, associated with expansion of the Lgr4+ve/ Lgr5+ve cells but also of Lgr4+ve/ Lgr5-ve cells [82]. In mammary glands, oncogenic mutation in the Lgr6+ve cells induces expansion of luminal cells generating tumor development [72]. In a mouse endometrial cancer model, Lgr5 is highly expressed in the initial stages of tumorigenesis but is downregulated in fully developed tumors [57]. In a skin carcinogenesis model involving UV or chemical treatments, Lgr5+ve or Lgr6+ve cells did not appear as tumor-initiating cells [83,84]. Conversely, in another study, oncogenic activation of b-catenin in Lgr5- or Lgr6-expressing cells of the skin lead to tumor development of different kinds [77]. Altogether, mouse models explored so far have generally recognized cells expressing the Lgrs as tumor-initiating cells. An overview of current knowledge on LGRs in human cancer is provided below.
LGRs alterations in cancer
Somatic mutations: Interrogation of the cBioPortal for cancer genomics database (http://www.cbioportal.org) indicates that genetic alterations are encountered for the LGR receptors in a large number of tumor types originating from bladder, breast, colorectal, esophageal, liver, lung, melanoma, pancreas, prostate, stomach and uterine tissues (Table 1). These genetic modifications can result from mutations, deletions, amplifications of multiple alterations. For example, mutations are often detected in colorectal, melanoma, lung, stomach or uterine tumors meanwhile amplifications seem to be frequent in prostate, pancreas and metastatic breast cancer. Further analysis of the genetic alterations detected in cancer tissues using the COSMIC database (http://cancer.sanger.ac.uk) shows that around 60% constitute missense substitutions that localize all along the coding sequence of LGR4, LGR5 and LGR6 (Figure 1B). With exception of the two colorectal cancer–associated LGR6 variants (G725C and P928H) mentioned earlier, the potential impact of such mutations on receptor activity and thereby on tumor initiation or maintenance still remains to be investigated but missense mutations may also simply represent passenger mutations. In addition, nonsense substitutions account for 4-8% of the reported genetic alterations. In particular for the LGR4 receptor, evidence has been provided that the nonsense c.376C>T mutation is associated with increased risk of squamous cell carcinoma and biliary tract cancer [85]. As the phenotype of human c.376C>T carriers is reminiscent of that exhibited by Lgr4-deficient mice, LGR4 has been proposed as a tumor suppressor gene [85].
|
|
|
LGR4 (% genetic alteration)
|
LGR5 (% genetic alteration)
|
LGR6 (% genetic alteration)
|
|
Tumor type
|
Reference study (retrieved from c-BioPortal)
|
Mutation
|
Deletion
|
Amplification
|
Multiple alterations
|
Mutation
|
Deletion
|
Amplification
|
Multiple alterations
|
Mutation
|
Deletion
|
Amplification
|
Multiple alterations
|
|
|
Bladder
|
Dana Farber & MSKCC, Cancer Discov 2014
|
4.00%
|
0.00%
|
0.00%
|
0.00%
|
2.00%
|
0.00%
|
0.00%
|
0.00%
|
not reported
|
|
|
|
TCGA Nature 2014
|
0.80%
|
0.80%
|
0.80%
|
0.00%
|
2.40%
|
0.00%
|
5.50%
|
0.00%
|
1.60%
|
0.80%
|
0.80%
|
0.00%
|
|
|
|
BGI, Nat Genet 2013
|
not reported
|
1.00%
|
0.00%
|
0.00%
|
0.00%
|
1.00%
|
0.00%
|
0.00%
|
0.00%
|
|
|
|
MSKCC, Eur Urol 2014
|
not reported
|
not reported
|
0.90%
|
0.00%
|
0.00%
|
0.00%
|
|
|
Breast
|
Igr France, 2016
|
0.00%
|
0.90%
|
3.30%
|
0.00%
|
1.40%
|
0.50%
|
8.00%
|
0.00%
|
0.00%
|
0.00%
|
6.60%
|
0.00%
|
|
|
|
British Columbia, Nature 2014
|
3.40%
|
0.00%
|
13.80%
|
0.00%
|
0.00%
|
0.00%
|
6.90%
|
0.00%
|
6.90%
|
0.00%
|
34.50%
|
0.00%
|
|
|
|
Broad, Nature 2012
|
1.00%
|
0.00%
|
0.00%
|
0.00%
|
not reported
|
not reported
|
|
|
|
METABRIC, Nature 2012 & Nat Commun 2016
|
0.00%
|
0.00%
|
1.80%
|
0.00%
|
0.00%
|
0.00%
|
2.50%
|
0.00%
|
0.00%
|
0.00%
|
24.40%
|
0.00%
|
|
|
|
Sanger, Nature 2012
|
4.00%
|
0.00%
|
0.00%
|
0.00%
|
not reported
|
not reported
|
|
|
|
TCGA, Provisional
|
0.40%
|
0.10%
|
0.90%
|
0.00%
|
0.70%
|
0.00%
|
2.70%
|
0.00%
|
0.20%
|
0.00%
|
11.60%
|
0.10%
|
|
|
Colorectal
|
DFCI, Cell Reports 2016
|
2.10%
|
0.00%
|
0.00%
|
0.00%
|
2.10%
|
0.00%
|
0.00%
|
0.00%
|
1.50%
|
0.00%
|
0.00%
|
0.00%
|
|
|
|
Genentech, Nature 2012
|
2.80%
|
0.00%
|
0.00%
|
0.00%
|
5.60%
|
0.00%
|
0.00%
|
0.00%
|
1.40%
|
0.00%
|
0.00%
|
0.00%
|
|
|
|
MSKCC, Genome Biol 2014
|
not reported
|
not reported
|
0.70%
|
0.00%
|
0.00%
|
0.00%
|
|
|
|
TCGA, Provisional
|
2.70%
|
0.50%
|
0.00%
|
0.00%
|
0.90%
|
0.00%
|
0.00%
|
0.00%
|
0.00%
|
0.00%
|
0.90%
|
0.00%
|
|
|
Esophageal adenocarcinoma
|
Broad, Nat Genet 2013
|
2.10%
|
0.00%
|
0.00%
|
0.00%
|
0%
|
0%
|
0%
|
0%
|
2.10%
|
0.00%
|
0.00%
|
0.00%
|
|
|
TCGA, Provisional
|
0.50%
|
0.00%
|
2.70%
|
0.00%
|
2.20%
|
0.00%
|
4.30%
|
0.00%
|
2.70%
|
0.00%
|
1.60%
|
0.00%
|
|
|
Esophageal squamous carcinoma
|
ICGC, Nature 2014
|
1.10%
|
0.00%
|
0.00%
|
0.00%
|
not reported
|
not reported
|
|
|
UCLA, Nat Genet 2014
|
not reported
|
not reported
|
2.20%
|
0.00%
|
0.00%
|
0.00%
|
|
|
Liver
|
AMC, Hepatology 2014
|
0.40%
|
0.00%
|
0.00%
|
0.00%
|
1.70%
|
0.00%
|
0.00%
|
0.00%
|
0.40%
|
0.00%
|
0.40%
|
0.00%
|
|
|
|
TCGA, Provisional
|
0.30%
|
0.00%
|
0.30%
|
0.00%
|
0.50%
|
0.00%
|
0.50%
|
0.00%
|
1.40%
|
0.00%
|
10.10%
|
0.00%
|
|
|
Lung adenocarcinoma
|
Broad, Cell 2012
|
3.30%
|
0.00%
|
0.50%
|
0.00%
|
1.60%
|
0.00%
|
2.70%
|
0.00%
|
3.30%
|
0.00%
|
2.70%
|
0.00%
|
|
|
TCGA, Nature 2014
|
0.40%
|
0.00%
|
1.70%
|
0.00%
|
4.80%
|
0.00%
|
3.50%
|
0.40%
|
2.60%
|
0.00%
|
7.40%
|
0.40%
|
|
|
MSKCC 2015
|
not reported
|
2.90%
|
0.00%
|
0.00%
|
0.00%
|
2.90%
|
0.00%
|
0.00%
|
0.00%
|
|
|
Pan-lung cancer
|
TCGA, Nat Genet 2016
|
2.00%
|
0.00%
|
0.70%
|
0.00%
|
3.30%
|
0.20%
|
2.40%
|
0.00%
|
3.00%
|
0.00%
|
3.10%
|
0.00%
|
|
|
Lung squamous carcinoma
|
TCGA, Nature 2012
|
1.70%
|
0.00%
|
0.00%
|
0.00%
|
1.70%
|
0.00%
|
1.70%
|
0.60%
|
1.10%
|
0.00%
|
0.00%
|
0.00%
|
|
|
Melanoma
|
Broad, Cell 2012
|
3.30%
|
0.00%
|
0.00%
|
0.00%
|
7.40%
|
0.00%
|
0.00%
|
0.00%
|
7.40%
|
0.00%
|
0.00%
|
0.00%
|
|
|
|
TCGA, Provisional
|
3.50%
|
0.00%
|
0.00%
|
0.00%
|
5.90%
|
0.00%
|
2.10%
|
0.30%
|
6.60%
|
0.00%
|
5.20%
|
0.00%
|
|
|
|
Yale, Nat Genet 2012
|
2.20%
|
0.00%
|
0.00%
|
0.00%
|
5.50%
|
0.00%
|
0.00%
|
0.00%
|
5.50%
|
0.00%
|
0.00%
|
0.00%
|
|
|
|
Broad/Dana Farber, Nature 2012
|
not reported
|
4.00%
|
0.00%
|
0.00%
|
0.00%
|
not reported
|
|
|
Pancreas
|
QCMG, Nature 2016
|
not reported
|
0.30%
|
0.00%
|
0.00%
|
0.00%
|
0.50%
|
0.00%
|
0.00%
|
0.00%
|
|
|
|
TCGA, Provisional
|
0.70%
|
0.00%
|
0.70%
|
0.00%
|
0.70%
|
0.00%
|
1.30%
|
0.00%
|
2.00%
|
0.00%
|
2.00%
|
0.00%
|
|
|
|
UTSW, Nat Commun 2015
|
0.00%
|
0.90%
|
7.30%
|
0.00%
|
0.90%
|
1.80%
|
3.70%
|
0.00%
|
0.00%
|
0.90%
|
5.50%
|
0.00%
|
|
|
|
Johns Hopkins University, Science 2011
|
not reported
|
10.00%
|
0.00%
|
0.00%
|
0.00%
|
not reported
|
|
|
Prostate
|
Broad/Cornell, Cell 2013
|
0.00%
|
0.00%
|
5.40%
|
0.00%
|
0.00%
|
1.60%
|
3.60%
|
0.00%
|
1.80%
|
0.00%
|
0.00%
|
0.00%
|
|
|
|
Fred Hutchinson CRC, Nat Med 2016
|
0.70%
|
0.00%
|
2.90%
|
0.00%
|
3.70%
|
0.00%
|
1.50%
|
0.00%
|
0.00%
|
0.00%
|
4.40%
|
0.00%
|
|
|
|
Michigan, Nature 2012
|
0.00%
|
0.00%
|
1.60%
|
0.00%
|
0.00%
|
0.00%
|
3.30%
|
0.00%
|
0.00%
|
0.00%
|
3.30%
|
0.00%
|
|
|
|
Robinson et al., Cell 2015
|
0.00%
|
0.00%
|
2.00%
|
0.00%
|
0.70%
|
0.00%
|
0.00%
|
0.00%
|
1.30%
|
0.00%
|
2.70%
|
0.00%
|
|
|
|
TCGA, Provisional
|
0.20%
|
0.00%
|
0.40%
|
0.00%
|
0.40%
|
0.20%
|
0.80%
|
0.00%
|
0.40%
|
0.40%
|
0.00%
|
0.00%
|
|
|
|
MSKCC, Cancer Cell 2010
|
not reported
|
0.00%
|
0.00%
|
1.00%
|
0.00%
|
not reported
|
|
|
Stomach
|
TCGA, Provisional
|
3.10%
|
0.30%
|
1.00%
|
0.00%
|
3.10%
|
0.30%
|
2.00%
|
0.00%
|
3.80%
|
0.30%
|
1.80%
|
0.00%
|
|
|
|
TMUCIH, PNAS 2015
|
3.80%
|
0.00%
|
0.00%
|
0.00%
|
not reported
|
not reported
|
|
|
|
UHK, Nat Genet 2011
|
13.60%
|
0.00%
|
0.00%
|
0.00%
|
not reported
|
4.50%
|
0.00%
|
0.00%
|
0.00%
|
|
|
|
U Tokyo, Nat Genet 2014
|
not reported
|
3.30%
|
0.00%
|
0.00%
|
0.00%
|
not reported
|
|
|
|
Pfizer and UHK, Nat Genet 2014
|
not reported
|
not reported
|
2.00%
|
0.00%
|
0.00%
|
0.00%
|
|
|
Uterine carcinosarcoma
|
Johns Hopkins University, Nat Commun 2014
|
4.50%
|
0.00%
|
0.00%
|
0.00%
|
not reported
|
4.50%
|
0.00%
|
0.00%
|
0.00%
|
|
|
TCGA, Provisional
|
1.80%
|
0.00%
|
1.80%
|
0.00%
|
5.40%
|
0.00%
|
3.60%
|
0.00%
|
3.60%
|
0.00%
|
5.40%
|
0.00%
|
|
|
Uterine Corpus Endometrial Carcinoma
|
TCGA, Provisional
|
3.70%
|
0.00%
|
0.80%
|
0.00%
|
2.10%
|
0.00%
|
2.10%
|
0.00%
|
4.10%
|
0.00%
|
3.30%
|
0.00%
|
|
Table 1. Genetic alteractions associated with LGR4, LGR5 and LGR6 in human cancer.
Splice variants: One relevant point is the potential differential function of splice variants on cell activity. Interrogation of the Ensembl database (http://www.ensembl.org) suggests the existence of 3 variants for LGR4 and LGR5 and 5 variants for LGR6, one of which is a transcript with nonsense-mediated decay. Interestingly, on soft-tissue sarcoma samples, two types of LGR5 transcripts has been detected, one encoding the full-length protein and one lacking the exon 5 (LGR5 Δex 5) that encodes part of the LRR4 till the LRR7 extracellular domain, largely encompassing the Rspondin–binding domain [86]. Low expression level of the shorter variant was correlated with later age of tumor onset but with poor prognosis for the disease-associated and recurrence-free survival, these data leading to propose LGR5 as an independent prognosis factor for soft-tissue sarcoma patients [86]. Recently, Lgr5 transcripts have been analyzed in intestinal tissue. In addition to the LGR5 full-length and Δex 5 transcripts, LGR5 variants missing exons 5 to 8 (Δex 5-8) or exon 8 (Δex 8) were detected [87]. The LGR5 full-length and splice variants were differentially expressed during cell cycle, being associated with cycle arrest and cycle progression, respectively. Moreover, they were associated with different proliferative capacity and sensitivity to chemotherapy [87]. In the paralogue LGR4, a naturally occurring splice variant has been identified in rat ovaries showing an antagonistic function in vitro on Wnt signaling as well as in vivo [88]. Whether such kind of LGR4 transcripts are also produced in cancer cells in variable proportion with respect to the full-length protein, which may alter its overall function has not been investigated.
Gene fusions: In addition to genetic alterations mentioned earlier, gene fusions are reported in the COSMIC and Atlas of Genetics and Cytogenetics in Oncology and Haematology databases (http://atlasgeneticsoncology.org/). Specifically, LGR4 fusions occur with CCDC34, NEMF, FGF3, DLGAP1 and TRIM58; LGR5 fusions occur with KIAA1033, TRHDE, INSR and NUP107, and LGR6 fusions occur with GPR37L1, PPP1R12B and RABGAP1L. So far, the potential impact of such alterations on LGRs and/or fusion partner function(s) is not known.
Indirect deregulation of LGR expression
Deregulation of the LGR expression may not be directly due to altered LGR sequence. This fact is exemplified by the LGR5 receptor, frequently overexpressed in cancer cells, and whose increased expression is most probably a consequence of the initial mutation in the Wnt signaling pathway; due to Apc loss-of-function or b-catenin gain-of-function [36]. Similarly, LGR4 expression is aberrantly induced by auto and/or paracrine IL6/JAK/Stat3 signaling in multiple myeloma cells [89]. In turn, stimulation of the receptor with Rspondin ligands locally produced by the niche (pre)osteoblasts results in Wnt co-receptors stabilization at the cell membrane and enhancement of the Wnt/b-catenin activity; which further sensitizes to auto- and paracrine Wnt ligands [89].
Epigenetics
Expression of the LGRs can also depend on the methylation status of their promoter region. An initial study reported that high expression of the Wnt-driven intestinal stem cells marker genes, including the receptor LGR5, was associated with a favorable prognosis among stage II colorectal cancer patients meanwhile LGR5 and other Wnt target genes became silenced by CpG island methylation during progression of tumorigenesis [90]. Another report on primary colorectal tumors has revealed that 40% of them exhibit LGR5 promoter methylation, leading to loss of receptor expression, a situation not detected in normal colon [91]. It is similarly suggested that silencing of LGR5 via CpG island methylation may be involved in disease progression, though in this case LGR5 methylation is rather proposed as a good prognosis marker [91]. Regarding the LGR6 receptor, its promoter has been reported to be hypermethylated in approximately 20% of sporadic colorectal cancer [92]. Interestingly, higher methylation level was detected in early as compared to advanced stages [92-94]. Together, these studies evidence that expression of LGR receptors is epigenetically modulated in cancer.
Prognostic value of LGRs
Expression of the LGR4 receptor promotes cancer cell proliferation in multiple cell types through activation of the Wnt signaling cascade [95,96]. In cervical carcinoma cells, it correlates with enhanced cell migration and metastasis [97]. In the case of skin and prostate, LGR4 promotes tumorigenesis by modulating MEK/ERK and Wnt/b-catenin pathways and the PI3K/Akt cascade, respectively [98,99]. In colorectal cancer, LGR4 expression is markedly upregulated in moderately and poorly differentiated cells as well as at the tumor invasive front and in metastasis and it significantly correlates with nodal spread in gastric cancer [100-102]. Altogether, studies point LGR4 as a poor-prognosis factor.
Much attention is currently focused on the LGR5 receptor due to its genuine association with stem cell identity. In ovarian, breast and lung cancer cells, high expression of LGR5 has been correlated with advanced stages, poor overall survival and metastasis [103-106]. In cervical cancer, LGR5 promotes proliferation and tumor formation through Wnt/b-catenin signaling [107]. LGR5 is reported as a cancer stem cell marker in gastric cancer, associated with stemness and EMT signature genes, and being positively correlated with well to moderate differentiation and nuclear b-catenin expression [108,109]. In line with these reports, a meta-analysis study on gastric cancer concludes that LGR5 overexpression is positively correlated with tumor stage, lymph node metastasis and poor overall survival [110]. In colorectal cancer, LGR5 is overexpressed in adenomas and frequently found in metastatic tissue, correlating with poor prognosis [103,111-114]. These reports are in line with two meta-analysis studies on colorectal cancer, in which LGR5 has been proposed as a poor prognosis marker [115,116]. Not yet explained is the clear controversy between multiple studies regarding the precise function of LGR5 (activator or negative regulator?) on the Wnt signaling cascade in cancer cells.
Regarding the paralogue LGR6, few studies are reported. In gastric cancer, its expression is significantly increased in tumors compared with corresponding non-neoplastic tissue and associated with local tumor growth [102]. Moreover, LGR6 expression has positive impact on patient survival in poorly cohesive-type carcinomas [102].
Conclusions and future directions
Since their initial discovery as close relatives of the glycoprotein hormone receptor family, LGR receptors have concentrated much attention in the stem cell and oncology research fields. The identification of LGRs, in particular LGR5, as stem-and cancer stem cell markers and evidences that these receptors can contribute to early stages of tumor development as well as to later metastatic processes, this provide rationale for proposing LGRs as interesting candidates for cancer treatments. One therapeutic approach currently being explored is to directly target the cancer cell using the LGR5 marker. Antibodies directed against the N-terminal domain of the LGR5 receptor, eliciting complement-dependent cytotoxicity or conjugated to a tubulin-inhibiting cytotoxic drug have been demonstrated to exhibit anti-tumor efficacy in xenograft models [117,118]. Further research will need to address how to circumvent the observed interconversion ability of cells to lose or re-express the LGR5 marker and to prevent tumor recurrence, knowing that both states sustain tumor-initiation activity [118-120]. Another approach would be to block the function of the LGR receptors in cancer cells by developing antibodies against the LGRs ligands or by producing soluble variants of the LGRs extracellular domains that would act as LGR antagonists. Moreover, there would be place for therapeutic strategies based on the design of drugs interfering with LGRs’ downstream signaling cascades (Wnt/bcatenin but also the MEK/Rho/ Wnt non-canonical pathways). Nevertheless, tremendous efforts are still needed in basic research to reach complete understanding on how LGRs exert their biological effects under homeostatic conditions as well as in cancer cells. Future studies aimed at dissecting the respective unique, redundant or opposing activities of each receptor on cells co-expressing LGRs will be of major relevance for the development of cancer treatments.
Authorship and contributorship
The review was written and edited by MIG.
Acknowledgments
We are grateful to Gilbert Vassart, Valeria Fernandez-Vallone and Romain Gerbier for critical reading of the manuscript.
Funding information
This work was supported by the Interuniversity Attraction Poles Programme- Belgian Science Policy (6/14), Belgian Fonds de la Recherche Scientifique Médicale, Walloon Region (CIBLES) and non-for-profit Association Recherche Biomédicale et Diagnostic.
References
- Hsu SY, Liang SG, Hsueh AJ (1998) Characterization of two LGR genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein-coupled, seven-transmembrane region. Mol Endocrinol 12: 1830-1845.
- Hsu SY, Kudo M, Chen T, Nakabayashi K, Bhalla A, et al. (2000) The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol Endocrinol 14: 1257-1271.
- Katritch V, Cherezov V, Stevens RC (2013) Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol 53: 531-556. [Crossref]
- Van Hiel MB, Vandersmissen HP, Van Loy T, Vanden Broeck J (2012) An evolutionary comparison of leucine-rich repeat containing G protein-coupled receptors reveals a novel LGR subtype. Peptides 34: 193-200.
- Xu JG, Huang C, Yang Z, Jin M, Fu P, et al. (2015) Crystal structure of LGR4-Rspo1 complex: insights into the divergent mechanisms of ligand recognition by leucine-rich repeat G-protein-coupled receptors (LGRs). J Biol Chem 290: 2455-2465.
- Vassart G, Pardo L, Costagliola S (2004) A molecular dissection of the glycoprotein hormone receptors. Trends Biochem Sci 29: 119-126. [Crossref]
- de Lau W, Barker N, Low TY, Koo BK, Li VS, et al. (2011) Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476: 293-297. [Crossref]
- Carmon KS, Gong X, Lin Q, Thomas A, Liu Q (2011) R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A 108: 11452-11457. [Crossref]
- Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, et al. (2011) LGR4 and LGR5 are R-spondin receptors mediating Wnt/b-catenin and Wnt/PCP signalling. EMBO Rep 12: 1055-1061. [Crossref]
- Ruffner H, Sprunger J, Charlat O, Leighton-Davies J, Grosshans B, et al. (2012) R-Spondin potentiates Wnt/b-catenin signaling through orphan receptors LGR4 and LGR5. PLoS One 7: e40976. [Crossref]
- Van Schoore G, Mendive F, Pochet R, Vassart G (2005) Expression pattern of the orphan receptor LGR4/GPR48 gene in the mouse. Histochem Cell Biol 124: 35-50. [Crossref]
- Mazerbourg S, Bouley DM, Sudo S, Klein CA, Zhang JV, et al. (2004) Leucine-rich repeat-containing, G protein-coupled receptor 4 null mice exhibit intrauterine growth retardation associated with embryonic and perinatal lethality. Mol Endocrinol 18: 2241-2254.
- Weng J, Luo J, Cheng X, Jin C, Zhou X, et al. (2008) Deletion of G protein-coupled receptor 48 leads to ocular anterior segment dysgenesis (ASD) through down-regulation of Pitx2. Proc Natl Acad Sci U S A 105: 6081-6086. [Crossref]
- Kato S, Mohri Y, Matsuo T, Ogawa E, Umezawa A, et al. (2007) Eye-open at birth phenotype with reduced keratinocyte motility in LGR4 null mice. FEBS Lett 581: 4685-4690. [Crossref]
- Mendive F, Laurent P, Van Schoore G, Skarnes W, Pochet R, et al. (2006) Defective postnatal development of the male reproductive tract in LGR4 knockout mice. Dev Biol 290: 421-434. [Crossref]
- Lambot MA, Mendive F, Laurent P, Van Schoore G, Noël JC, et al. (2009) Three-dimensional reconstruction of efferent ducts in wild-type and Lgr4 knock-out mice. Anat Rec (Hoboken) 292: 595-603. [Crossref]
- Kato S, Matsubara M, Matsuo T, Mohri Y, Kazama I, et al. (2006) Leucine-rich repeat-containing G protein-coupled receptor-4 (LGR4, Gpr48) is essential for renal development in mice. Nephron Exp Nephrol 104: e63-75. [Crossref]
- Yamashita R, Takegawa Y, Sakumoto M, Nakahara M, Kawazu H, et al. (2009) Defective development of the gall bladder and cystic duct in Lgr4- hypomorphic mice. Dev Dyn 238: 993-1000. [Crossref]
- Mohri Y, Kato S, Umezawa A, Okuyama R, Nishimori K (2008) Impaired hair placode formation with reduced expression of hair follicle-related genes in mice lacking Lgr4. Dev Dyn 237, 2235–2242.
- Mohri Y, Umezu T, Hidema S, Tomisawa H, Akamatsu A, et al. (2010) Reduced fertility with impairment of early-stage embryos observed in mice lacking Lgr4 in epithelial tissues. Fertil Steril 94: 2878-2881.
- Mohri Y, Oyama K, Akamatsu A, Kato S, Nishimori K (2011) Lgr4-deficient mice showed premature differentiation of ureteric bud with reduced expression of Wnt effector Lef1 and Gata3. Dev Dyn 240: 1626-1634.
- Sone M, Oyama K, Mohri Y, Hayashi R, Clevers H, et al. (2013) LGR4 expressed in uterine epithelium is necessary for uterine gland development and contributes to decidualization in mice. FASEB J 27: 4917-4928. [Crossref]
- Yamakami Y, Kohashi K, Oyama K, Mohri Y, Hidema S, et al. (2016) LGR4 is required for sequential molar development. Biochem Biophys Rep 8: 174-183.
- Kida T, Oyama K, Sone M, Koizumi M, Hidema S, et al. (2014) Lgr4 is required for endometrial receptivity acquired through ovarian hormone signaling. Biosci Biotechnol Biochem 78: 1813-1816.
- Oyama K, Mohri Y, Sone M, Nawa A, Nishimori K (2011) Conditional knockout of Lgr4 leads to impaired ductal elongation and branching morphogenesis in mouse mammary glands. Sex Dev 5: 205-212.
- Wang Z, Jin C, Li H, Li C, Hou Q, et al. (2010) GPR48-Induced keratinocyte proliferation occurs through HB-EGF mediated EGFR transactivation. FEBS Lett 584: 4057-4062. [Crossref]
- Song H, Luo J, Luo W, Weng J, Wang Z, et al. (2008) Inactivation of G-protein-coupled receptor 48 (Gpr48/Lgr4) impairs definitive erythropoiesis at midgestation through down-regulation of the ATF4 signaling pathway. J Biol Chem 283: 36687-36697. [Crossref]
- Mustata RC, Van Loy T, Lefort A, Libert F, Strollo S, et al. (2011) Lgr4 is required for Paneth cell differentiation and maintenance of intestinal stem cells ex vivo. EMBO Rep 12: 558-564. [Crossref]
2021 Copyright OAT. All rights reserv
- Kinzel B, Pikiolek M1, Orsini V1, Sprunger J1, Isken A1, et al. (2014) Functional roles of Lgr4 and Lgr5 in embryonic gut, kidney and skin development in mice. Dev Biol 390: 181-190. [Crossref]
- Zak M, van Oort T, Hendriksen FG, Garcia MI, Vassart G, et al. (2016) LGR4 and LGR5 Regulate Hair Cell Differentiation in the Sensory Epithelium of the Developing Mouse Cochlea. Front Cell Neurosci 10: 186. [Crossref]
- Qian Y, Liu S, Guan Y, Pan H, Guan X, et al. (2013) Lgr4-mediated Wnt/β-catenin signaling in peritubular myoid cells is essential for spermatogenesis. Development 140: 1751-1761. [Crossref]
- Wang J, Li X, Ke Y, Lu Y, Wang F, et al. (2012) GPR48 increases mineralocorticoid receptor gene expression. J Am Soc Nephrol 23: 281-293. [Crossref]
- Luo J, Zhou W, Zhou X, Li D, Weng J, et al. (2009) Regulation of bone formation and remodeling by G-protein-coupled receptor 48. Development 136: 2747-2756. [Crossref]
- Liu S, Qian Y, Li L, Wei G, Guan Y, et al. (2013) Lgr4 gene deficiency increases susceptibility and severity of dextran sodium sulfate-induced inflammatory bowel disease in mice. J Biol Chem 288: 8794-8803.
- Planas-Paz L, Orsini V, Boulter L, Calabrese D, Pikiolek M, et al. (2016) The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat Cell Biol 18: 467-479.
- Zhan T, Rindtorff N, Boutros M (2017) Wnt signaling in cancer. Oncogene 36: 1461-1473. [Crossref]
- Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, et al. (2012) ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485: 195-200. [Crossref]
- Koo BK, Spit M, Jordens I, Low TY, Stange DE, et al. (2012) Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488: 665-669. [Crossref]
- Carmon KS, Gong X, Yi J, Thomas A, Liu Q (2014) RSPO-LGR4 functions via IQGAP1 to potentiate Wnt signaling. Proc Natl Acad Sci U S A 111: E1221-1229. [Crossref]
- Deng C, Reddy P, Cheng Y, Luo CW, Hsiao CL, et al. (2013) Multi-functional norrin is a ligand for the LGR4 receptor. J Cell Sci 126: 2060-2068. [Crossref]
- Luo J, Yang Z, Ma Y, Yue Z, Lin H, et al. (2016) LGR4 is a receptor for RANKL and negatively regulates osteoclast differentiation and bone resorption. Nat Med 22: 539-546. [Crossref]
- Zhu C, Zheng XF, Yang YH, Li B, Wang YR, et al. (2016) LGR4 acts as a key receptor for R-spondin 2 to promote osteogenesis through Wnt signaling pathway. Cell Signal 28: 989-1000. [Crossref]
- Wang J, Liu R, Wang F, Hong J, Li X, et al. (2013) Ablation of LGR4 promotes energy expenditure by driving white-to-brown fat switch. Nat Cell Biol 15: 1455-1463. [Crossref]
- Zou Y, Ning T, Shi J, Chen M, Ding L, et al. (2017) Association of a gain-of-function variant in LGR4 with central obesity. Obesity (Silver Spring) 25: 252-260. [Crossref]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, et al. (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003-1007. [Crossref]
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, et al. (2010) Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6: 25-36. [Crossref]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, et al. (2008) Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet 40: 1291-1299. [Crossref]
- Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJ, et al. (2013) Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 32: 2708-2721. [Crossref]
- Barker N, Rookmaaker MB, Kujala P, Ng A, Leushacke M, et al. (2012) Lgr5(+ve) stem/progenitor cells contribute to nephron formation during kidney development. Cell Rep 2: 540-552. [Crossref]
- Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, et al. (2013) Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep 3: 70-78. [Crossref]
- Visser KE, Ciampricotti M, Michalak EM, Tan DW, Speksnijder EN, et al. (2012) Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol 228: 300-309.
- Lehoczky JA, Tabin CJ (2015) Lgr6 marks nail stem cells and is required for digit tip regeneration. Proc Natl Acad Sci U S A 112: 13249-13254. [Crossref]
- Ren W, Lewandowski BC, Watson J, Aihara E, Iwatsuki K, et al. (2014) Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc Natl Acad Sci U S A 111: 16401-16406. [Crossref]
- Morita H, Mazerbourg S, Bouley DM, Luo CW, Kawamura K, et al. (2004) Neonatal lethality of LGR5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol Cell Biol 24: 9736-9743. [Crossref]
- Garcia MI, Ghiani M, Lefort A, Libert F, Strollo S, et al. (2009) LGR5 deficiency deregulates Wnt signaling and leads to precocious Paneth cell differentiation in the fetal intestine. Dev Biol 331: 58-67. [Crossref]
- Walker F, Zhang HH, Odorizzi A, Burgess AW (2011) LGR5 is a negative regulator of tumourigenicity, antagonizes Wnt signalling and regulates cell adhesion in colorectal cancer cell lines. PloS One 6: e22733.
- Sun X, Jackson L, Dey SK, Daikoku T (2009) In pursuit of leucine-rich repeat-containing G protein-coupled receptor-5 regulation and function in the uterus. Endocrinology 150: 5065-5073. [Crossref]
- Lewis A, Segditsas S, Deheragoda M, Pollard P, Jeffery R, et al. (2010) Severe polyposis in Apc(1322T) mice is associated with submaximal Wnt signalling and increased expression of the stem cell marker Lgr5. Gut 59: 1680-1686. [Crossref]
- Xu K, Xu Y, Rajashankar KR, Robev D, Nikolov DB (2013) Crystal structures of Lgr4 and its complex with R-spondin1. Structure 21: 1683-1689. [Crossref]
- Peng WC, de Lau W, Forneris F, Granneman JC, Huch M, et al. (2013) Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell Rep 3: 1885-1892. [Crossref]
- Carmon KS, Lin Q, Gong X, Thomas A, Liu Q (2012) LGR5 interacts and cointernalizes with Wnt receptors to modulate Wnt/b-catenin signaling. Mol Cell Biol 32: 2054-2064. [Crossref]
- Hirata‐Tominaga K, Nakamura T, Okumura N, Kawasaki S, Kay EP, et al. (2013) Corneal endothelial cell fate is maintained by LGR5 through the regulation of hedgehog and Wnt pathway. Stem Cells 31: 1396-1407.
- Wu C, Qiu S, Lu L, Zou J, Li WF, et al. (2014) RSPO2-LGR5 signaling has tumour-suppressive activity in colorectal cancer. Nat Commun 5: 3149. [Crossref]
- Fafilek B, Krausova M, Vojtechova M, Pospichalova V, Tumova L, et al. (2013) Troy, a tumor necrosis factor receptor family member, interacts with lgr5 to inhibit wnt signaling in intestinal stem cells. Gastroenterol 144: 381-391.
- Snyder JC, Rochelle LK, Lyerly HK, Caron MG, Barak LS (2013) Constitutive internalization of the leucine-rich G protein-coupled receptor-5 (LGR5) to the trans-Golgi network. J Biol Chem 288: 10286-10297.
- Snyder JC, Rochelle LK, Barak LS, Caron MG (2013) The stem cell-expressed receptor Lgr5 possesses canonical and functionally active molecular determinants critical to ß-arrestin-2 recruitment. PloS One 8: e84476.
- Snyder JC, Rochelle LK, Ray C, Pack TF, Bock CB, et al. (2017) Inhibiting clathrin-mediated endocytosis of the leucine-rich G protein-coupled Receptor-5 diminishes cell fitness. J Biol Chem. [Crossref]
- Mueller S, Jaeschke H, Günther R, Paschke R (2010) The hinge region: an important receptor component for GPHR function. Trends Endocrinol Metab 21: 111-122.
- Shaikh LH, Zhou J, Teo AE, Garg S, Neogi SG, et al. (2015) LGR5 Activates Noncanonical Wnt Signaling and Inhibits Aldosterone Production in the Human Adrenal. J Clin Endocrinol Metab 100: E836-844. [Crossref]
- Kwon MS, Park BO, Kim HM, Kim S (2013) Leucine-rich repeat-containing G-protein coupled receptor 5/GPR49 activates G12/13-Rho GTPase pathway. Mol Cells 36: 267-272. [Crossref]
- Zhang Y, Chen Y, Ni W, Guo L, Lu X, et al. (2015) Dynamic expression of Lgr6 in the developing and mature mouse cochlea. Front Cell Neurosci 9: 165. [Crossref]
- Blaas L, Pucci F, Messal HA, Andersson AB, Ruiz EJ, et al. (2016) Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat Cell Biol 18: 1346-1356.
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, et al. (2010) Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327: 1385-1389. [Crossref]
- Liao XH, Nguyen H (2014) Epidermal expression of Lgr6 is dependent on nerve endings and Schwann cells. Exp Dermatol 23: 195-198. [Crossref]
- Füllgrabe A, Joost S, Are A, Jacob T, Sivan U, et al. (2015) Dynamics of Lgr6+ Progenitor Cells in the Hair Follicle, Sebaceous Gland, and Interfollicular Epidermis. Stem Cell Reports 5: 843-855. [Crossref]
- Jiang L, Wang L, Chen CP, Li MQ, Liao XH (2017) Lgr6 is dispensable for epidermal cell proliferation and wound repair. Exp Dermatol 26: 105-107. [Crossref]
- Kretzschmar K, Weber C, Driskell RR, Calonje E, Watt FM (2016) Compartmentalized Epidermal Activation of ß-Catenin Differentially Affects Lineage Reprogramming and Underlies Tumor Heterogeneity. Cell Rep 14: 269-281.
- Oeztuerk-Winder F, Guinot A, Ochalek A, Ventura JJ (2012) Regulation of human lung alveolar multipotent cells by a novel p38α MAPK/miR-17-92 axis. EMBO J 31: 3431-3441. [Crossref]
- Gong X, Carmon KS, Lin Q, Thomas A, Yi J, et al. (2012) LGR6 is a high affinity receptor of R-spondins and potentially functions as a tumor suppressor. PLoS One 7: e37137. [Crossref]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, et al. (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457: 608-611. [Crossref]
- Yanai H, Atsumi N, Tanaka T, Nakamura N, Komai Y, et al. (2017) Intestinal cancer stem cells marked by Bmi1 or Lgr5 expression contribute to tumor propagation via clonal expansion. Sci Rep 7: 41838. [Crossref]
- Hilkens J, Timmer NC, Boer M, Ikink GJ, Schewe M, et al. (2016) RSPO3 expands intestinal stem cell and niche compartments and drives tumorigenesis. Gut. [Crossref]
- van de Glind GC, Out JJ, Rebel HG, Tensen CP, de Gruijl FR (2016) Lgr6+ stem cells and their progeny in mouse epidermis under regimens of exogenous skin carcinogenesis, and their absence in ensuing skin tumors. Oncotarget 7: 86740-86754.
- van de Glind GC, Out JJ, Rebel HG, Tensen CP, de Gruijl FR (2016) Lgr5+ stem cells and their progeny in mouse epidermis under regimens of exogenous skin carcinogenesis, and their absence in ensuing skin tumors. Oncotarget 7: 52085-52094.
- Styrkarsdottir U, Thorleifsson G, Sulem P, Gudbjartsson DF, Sigurdsson A, et al. (2013) Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature 497: 517-520. [Crossref]
- Rot S, Taubert H, Bache M, Greither T, Würl P, et al. (2011) A novel splice variant of the stem cell marker LGR5/GPR49 is correlated with the risk of tumor-related death in soft-tissue sarcoma patients. BMC Cancer 11: 429. [Crossref]
- Osawa H, Takahashi H, Nishimura J, Ohta K, Haraguchi N, et al. (2016) Full-length LGR5-positive cells have chemoresistant characteristics in colorectal cancer. Br J Cancer 114: 1251-1260. [Crossref]
- Hsu PJ, Wu FJ, Kudo M, Hsiao CL, Hsueh AJ, et al. (2014) A naturally occurring Lgr4 splice variant encodes a soluble antagonist useful for demonstrating the gonadal roles of Lgr4 in mammals. PloS One 9: e106804.
- van Andel H, Ren Z, Koopmans I, Joosten SP, Kocemba KA, et al. (2017) Aberrantly expressed LGR4 empowers Wnt signaling in multiple myeloma by hijacking osteoblast-derived R-spondins. Proc Natl Acad Sci U S A 114: 376-381. [Crossref]
- de Sousa E Melo F, Colak S, Buikhuisen J, Koster J, Cameron K, et al. (2011) Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 9: 476-485. [Crossref]
- Su S, Hong F, Liang Y, Zhou J, Liang Y, et al. (2015) Lgr5 Methylation in Cancer Stem Cell Differentiation and Prognosis-Prediction in Colorectal Cancer. PLoS One 10: e0143513. [Crossref]
- Mokarram P, Kumar K, Brim H, Naghibalhossaini F, Saberi-firoozi M, et al. (2009) Distinct high-profile methylated genes in colorectal cancer. PLoS One 4: e7012. [Crossref]
- Brim H, Abu-Asab MS, Nouraie M, Salazar J, Deleo J, et al. (2014) An integrative CGH, MSI and candidate genes methylation analysis of colorectal tumors. PLoS One 9: e82185. [Crossref]
- Chan TA, Glockner S, Yi JM, Chen W, Van Neste L, et al. (2008) Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med 5: e114. [Crossref]
- Zhu YB, Xu L, Chen M, Ma HN, Lou F (2013) GPR48 promotes multiple cancer cell proliferation via activation of Wnt signaling. Asian Pac J Cancer Prev 14: 4775-4778. [Crossref]
- Yu CY, Liang GB, Du P, Liu YH (2013) Lgr4 promotes glioma cell proliferation through activation of Wnt signaling. Asian Pac J Cancer Prev 14: 4907-4911. [Crossref]
- Gao Y, Shan ZY, Wang H, Zhang HM, Teng WP (2009) Inhibitory effect of shRNA targeting GPR48 on invasion and metastasis of human cervical carcinoma cell line HeLa. Ai Zheng 28: 104-107.
- Xu P, Dang Y, Wang L, Liu X, Ren X, et al. (2016) Lgr4 is crucial for skin carcinogenesis by regulating MEK/ERK and Wnt/b-catenin signaling pathways. Cancer Lett 383: 161-170. [Crossref]
- Liang F, Yue J, Wang J, Zhang L, Fan R, et al. (2015) GPCR48/LGR4 promotes tumorigenesis of prostate cancer via PI3K/Akt signaling pathway. Med Oncol 32: 49. [Crossref]
- Gao Y, Kitagawa K, Hiramatsu Y, Kikuchi H, Isobe T, et al. (2006) Up-regulation of GPR48 induced by down-regulation of p27Kip1 enhances carcinoma cell invasiveness and metastasis. Cancer Res 66: 11623-11631. [Crossref]
- Wu J, Xie N, Xie K, Zeng J, Cheng L, et al. (2013) GPR48, a poor prognostic factor, promotes tumor metastasis and activates b-catenin/TCF signaling in colorectal cancer. Carcinogenesis 34: 2861-2869. [Crossref]
- Steffen JS, Simon E, Warneke V, Balschun K, Ebert M, et al. (2012) LGR4 and LGR6 are differentially expressed and of putative tumor biological significance in gastric carcinoma. Virchows Arch 461: 355-365.
- McClanahan T, Koseoglu S, Smith K, Grein J, Gustafson E, et al. (2006) Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol Ther 5: 419-426. [Crossref]
- Sun Y, Jia X, Wu X (2015) High Expressions of Lgr5 and ALDH1 in Primary Epithelial Ovarian Cancer Correlate with Advanced Tumor Stage and Grade as well as Poor Prognosis of the Patients. Gynecol Obstet Invest 81: 162-168.
- Yang L, Tang H, Kong Y, Xie X, Chen J, et al. (2015) LGR5 Promotes Breast Cancer Progression and Maintains Stem-Like Cells Through Activation of Wnt/b-catenin Signaling. Stem Cells 33: 2913-2924. [Crossref]
- Zhang X, Xu M, Su S, Zhou Z, Yang H, et al. (2016) Lgr5-positive cells in the lung and their clinical significance in patients with lung adenocarcinoma. Mol Clin Oncol 5: 283-288. [Crossref]
- Chen Q, Cao HZ, Zheng PS (2014) LGR5 promotes the proliferation and tumor formation of cervical cancer cells through the Wnt/ß-catenin signaling pathway. Oncotarget 5: 9092-9105.
- Wang B, Chen Q, Cao Y, Ma X, Yin C, et al. (2016) LGR5 Is a Gastric Cancer Stem Cell Marker Associated with Stemness and the EMT Signature Genes NANOG, NANOGP8, PRRX1, TWIST1, and BMI1. PLoS One 11: e0168904. [Crossref]
- Jang BG, Lee BL, Kim WH (2016) Prognostic significance of leucine-rich-repeat-containing G-protein-coupled receptor 5, an intestinal stem cell marker, in gastric carcinomas. Gastric Cancer 19: 767-777.
- Huang T, Qiu X, Xiao J, Wang Q, Wang Y, et al. (2016) The prognostic role of Leucine-rich repeat-containing G-protein-coupled receptor 5 in gastric cancer: A systematic review with meta-analysis. Clin Res Hepatol Gastroenterol 40: 246-253. [Crossref]
- Baker AM, Graham TA, Elia G, Wright NA, Rodriguez-Justo M (2015) Characterization of LGR5 stem cells in colorectal adenomas and carcinomas. Sci Rep 5: 8654. [Crossref]
- Uchida H, Yamazaki K, Fukuma M, Yamada T, Hayashida T, et al. (2010) Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in colorectal cancer. Cancer Sci 101: 1731-1737. [Crossref]
- Gao FJ, Chen JY, Wu HY, Shi J, Chen M, et al. (2014) Lgr5 over-expression is positively related to the tumor progression and HER2 expression in stage pTNM IV colorectal cancer. Int J Clin Exp Pathol 7: 1572-1579. [Crossref]
- Wu W, Cao J, Ji Z, Wang J, Jiang T, et al. (2016) Co-expression of Lgr5 and CXCR4 characterizes cancer stem-like cells of colorectal cancer. Oncotarget 7: 81144-81155. [Crossref]
- Han Y, Xue X, Jiang M, Guo X, Li P, et al. (2015) LGR5, a relevant marker of cancer stem cells, indicates a poor prognosis in colorectal cancer patients: a meta-analysis. Clin Res Hepatol Gastroenterol 39: 267-273.
- Jiang Y, Li W, He X, Zhang H, Jiang F, et al. (2015) Lgr5 expression is a valuable prognostic factor for colorectal cancer: evidence from a meta-analysis. BMC Cancer 15: 948.
- Sasaki Y, Kosaka H, Usami K, Toki H, Kawai H, et al. (2010) Establishment of a novel monoclonal antibody against LGR5. Biochem Biophys Res Commun 394: 498-502. [Crossref]
- Gong X, Azhdarinia A, Ghosh SC, Xiong W, An Z, et al. (2016) LGR5-Targeted Antibody-Drug Conjugate Eradicates Gastrointestinal Tumors and Prevents Recurrence. Mol Cancer Ther 15: 1580-1590.
- Kobayashi S, Yamada‐Okabe H, Suzuki M, Natori O, Kato A, et al. (2012) LGR5-positive colon cancer stem cells interconvert with drug-resistant LGR5-negative cells and are capable of tumor reconstitution. Stem Cells 30: 2631-2644.
- Melo FS, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, et al. (2017) A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543: 676-680. [Crossref]

