FAM20A is a member of family of related proteins that has been named family with sequence similarity 20 (FAM20) with three members (FAM20A, FAM20B and FAM20C) in mammals. Recently, some studies have reported that patients with
Amelogenesis Imperfecta
caused by FAM20A mutation displayed several dental phenotypes including hypoplastic enamel, failure of tooth development and gingival hyperplasia. We have reported a transgenic mouse line with a 58-kb fragment deletion that encompasses part of the Fam20a gene and its upstream region shows growth disorder. In the present study, we aimed to clarify how the loss of Fam20a protein influenced the development of dental tissues in the
Fam20a-/-mouse. The light microscopic study (LM) of paraffin-embedded specimens demonstrated hypoplastic enamel and gingival hyperplasia in the Fam20a-/-mice. The LM of ground sections of the mandibles containing teeth showed that there was no enamel covering the coronal dentine. Immunohistochemical study of the mandibles (paraffin-embedded specimens) indicated that Fam20a protein expressed in osteoid tissue and tooth germ of Fam20a+/+mice, but does not expressed in Fam20a-/- mice. In conclusion, this study revealed that the FAM20A protein is required for enamel formation.
amelogenesis imperfecta, gingival hyperplasia, fam20a
Amelogenesis imperfecta (AI) is a collective group of inherited enamel defects without any normal syndromic phenotypes [1]. On the other hand, a number of inherited congenital defects of enamel formation have been described, together termed AI, that provide the opportunity to delineate the essential events underlying biomineralization and to dissect the molecular pathogenesis of skeletal disease [2]. Dental enamel is the most highly mineralized tissue in the human body. Mature enamel contains more than 95% mineral content while its organic content is less than 1% [3]. A series of ecto-mesenchymal interactions needs to be well coordinated in the development of sound teeth, and the initial secretion of the dentin matrix by the odontoblasts is followed by the secretion of the enamel matrix by the ameloblasts [4]. The secreted enamel matrix is then calcified and subsequently matured by the removal of the remaining enamel matrix proteins and by the growth of the enamel crystallites. Alterations in any of these steps induce enamel defects. A reduction in the secretion of the enamel matrix causes hypoplastic AI, in which the enamel is thin. A previous study has observed that hypocalcified enamel was extremely soft but the thickness of the enamel was normal. In contrast, hypomaturated enamel was soft and discolored, but had a normal thickness [5].
Family with sequence similarity 20 (FAM20) consists of three members (FAM20A, FAM20B, and FAM20C) and it has been suggested that these genes evolved from a common ancestral gene through a gene duplication event [6]. Fam20a gene is located on chromosome 11E1 and consists of 11 exons spanning around 60 kb. Translation starts in the first exon, terminates in the last exon, and encodes a protein of 541 amino acids, which has been shown to be a secreted glycoprotein [6]. The FAM20 family of secreted proteins in mammals consists of three members (FAM20A, FAM20B, and FAM20C) that were initially thought to have roles in regulating the differentiation and function of hematopoietic and other tissues [6]. In humans, a mutation in FAM20A was recently linked to the occurrence of AI and gingival hyperplasia in a consanguineous family [7].
Recently, we have reported our discovery of a transgenic (Tg) mouse line exhibiting growth disorder, which we discovered in the course of generating Tg mice expressing a viral gene [8]. These mice did not express any corresponding viral mRNA or protein due to a deletion in the transgene, with a 58-kb fragment deletion in chromosome 11E1 that encompasses part of the Fam20a gene [8]. In the present study, we reported AI observed in a Fam20a Knockout mice.
FAM20A Knockout mice
We have previously reported a Tg mouse lineage (line 230) with growth disorder and the transgene mRNA expression was not detected in the liver [8]. Nevertheless,the transgene insertion resulted in the deletion of a 58-kb genome fragment encompassing part of the Fam20a gene, including exon 1, and its upstream region. RT-PCR analysis suggest that the homozygous progeny of line 230 Tg mice not expressing Fam20a mRNA(Figure 1). We belived that this line230 Tg mice equivalent with FAM20A Knockout mice.
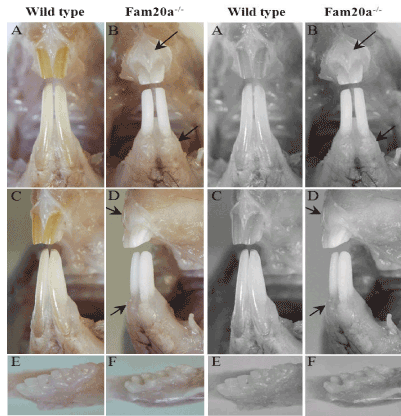
Figure 1. Visual examination
Incisor teeth; Fam20a+/+ mouse: The enamel of incisors in wild-type mice typically shows a smooth opalescent surface and has yellow brown coloration due to iron content (A, C). Incisor teeth; Fam20a–/– mouse: Both the upper and lower incisors in Fam20a–/– mice had a chalky white to opaque appearance (B, D). Mandibles with molar teeth; Fam20 +/+ mouse: The molar teeth in wild-type mice have creamy white, translucent enamel and smooth surfaces (E). Mandibles with molar teeth; Fam20–/–mouse: Whereas the molar teeth in Fam20a–/– mice has white, opaque enamel showed roughened surfaces (F). Gingiva; Fam20a–/– mouse: Compare with wild-type mice, Fam20a–/– mice showed significant enlarged gingiva (black arrow).
Mouse husbandry and testing
Mice were cared for according to institutional guidelines, fed with ordinary feed (Funashi Farms, Funabashi, Japan) and maintained in specific pathogen-free conditions. All of the animal experiments were carried out according to the protocol approved by the Ethics Committee for Animal Experiments at Kobe University.
Quarterly sentinel surveillance showed no evidence of pathogenic rodent viruses, Mycoplasma, or Helicobacter species in the Lexicon Pharmaceutical source colonies.
In this study, observations were normally completed in mice ranging in age from 8 to 60 weeks. Statistical comparisons were made using the 2-tail Student’s t-test, based on inspection of group data and the assumption that the group data show a normal distribution.
Analysis of Fam20a mRNA expression
Total RNA was extracted from the liver with an RNeasy Mini Kit (QIAGEN). A hundred ng total RNA was reverse-transcribed and amplified using SuperScript One-step RT-PCR (Invitrogen). The following primers were used for the RT-PCR: H5-5 38-F (5’-TACTCGGTTGCATCGTCACT-3’) and H5-5 335-R (5’-TCAGCATGTCTCGTGACCAA-3’) for mRNA of Fam20a.
Histopathological analysis and Immuno-fluorescence-based staining
Tissue specimens (head portions of mice) were fixed in 4% paraformaldehyde, dehydrated in an increasing sequence of ethyl alcohol solutions, and embedded in paraffin. The specimens were then sectioned at a thickness of 5 µm and stained with hematoxylin-eosin (HE).The expression and localization of Fam20a protein were examined using anti-Fam20a antibody by immuno-fluorescence-based staining.
Ground sections of the mandible
Mandibles containing incisor and molars were dissected from the head portions, ground-sections (30 m) after removal of soft tissues, treated with HCl (0.3 N, 2 seconds), stained with carbolfuchsin (0.1 w/v %) and prepared for the present LM of hard tissues.
Visual examination
Although the number of homozygous [HOM] mice was significantly lower than expected, Fam20a–/– mice were produced in approximately
(equal
) 1/2 numbers of Fam20a+/+ mice (WT: 52; heterozygous [HET]: 96; HOM: 27). Fam20a+/+ or Fam20a–/+ mice displayed essentially normal
growth rate
2021 Copyright OAT. All rights reserv
s,
growth rates of Fam20a–/– mice
decreased 50% and every type mice
(Fam20a+/+
, Fam20a–/+,
Fam20a–/–) may survive up to one year of age. Fertility of both males and females
in the Fam20a+/+ and Fam20a–/+ mice was also normal. In the Fam20a–/– mice, estrus is usually two weeks late than normal mice and farrowing rate reduced by 50%. The most striking gross lesions were noted in the dentition of Fam20a–/– mice. Initial pathological evaluations were completed on 6 Fam20a–/– mice (2 male and 4 female) aged between 10 and 12 weeks, while a second cohort of 6 Fam20a–/– mice (2 male and 4 female) was necropsied between 54 and 60 weeks of age.
Normal incisors in wild-type and heterozygous mice typically show yellow brown coloration with a smooth opalescent surface (Figure 1A and C). In contrast, both the upper and lower incisors in Fam20a–/– mice had a chalky white to opaque appearance (Figure 1B and D). The molar teeth of young Fam20a–/– mice were small and peg-like (Figure 1F), with an opaque and roughened surface, whereas the molar teeth of wild-type and heterozygous littermate mice were smooth and opalescent (Figure 1E). In the older Fam20a–/– mice, the gross appearance of incisor teeth was essentially unchanged, but the molar teeth had worn down. Obvious enlarged gingiva was observed in Fam20a–/– mouse compare with that of wild-type mice.
LM study
Histologically, the normal developing incisor clearly show enamel, dentin and cementum in the ground sections
(Figure 2A)
, and the enamel consists of highly organized arrangements of decussating enamel rods (Figure 2C). We found that tooth lesions in Fam20a–/– mice were essentially restricted to the enamel layer, with all incisor and molar teeth lacking true enamel (Figure 2B and D). No abnormalities were detected in the dentin and cementum.
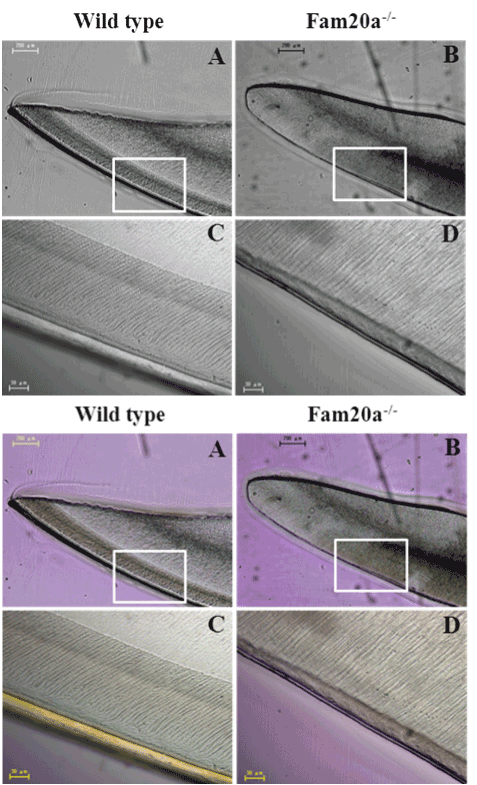
Figure 2. LM study.
Incisor tooth; Fam20a+/+ mouse: Regularly-arranged prismatic structures were observed in the enamel of wild type mice. Ground sections. (A, C). incisor tooth; Fam20a–/– mouse: no prismatic structures were demonstrated in the superficial layer covering the labial dentine of Fam20a–/– mice (B, D).
X-ray examination
X-ray radiography of the wild type and FAM20A-/- mice showing the dimensions of the Fam20a–/– mouse skull were smaller than those of the wild type mice (i.e., width: 102 to 100, length: 93 to 100) (Figure 3A and D). The length of the Fam20a–/– mouse molar region was about 85% of that of wild type mice (black arrow; Figure 3B and E). The tooth thickness (labio-lingual dimension) of the Fam20a–/– mouse lower incisor was about 72% of that of wild type mice (black arrow; Figure 3C and F), while there was no significant differences in the internal diameter of alveolus (white arrow; Figure 3C and F).
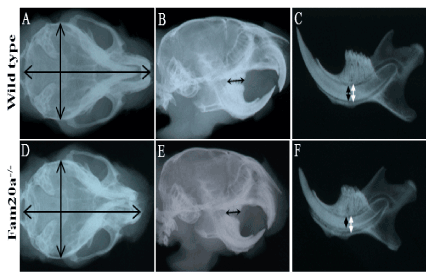
Figure 3. X-ray examination.
Skull size; Fam20a+/+ and Fam20a–/– mouse: The dimensions of the Fam20a–/– mouse skull were smaller than those of the wild type mice. X-ray radiography. (i.e., width: 102 to 100, length: 93 to 100). (A, D). Molar region; Fam20a+/+ and Fam20a–/– mouse: The length of the Fam20a–/– mouse molar region was about 85% of that of wild type mice. X-ray radiography. (B, E. black arrow). Incisor tooth; Fam20a+/+ and Fam20a–/– mouse: the thickness of the Fam20a–/– mouse lower incisor was about 72% of that of wild type mice (C, F. black arrow), while there was no significant differences in the internal diameter of alveolus (C, F. white arrow). X-ray radiography.
Immunohistochemistry
Immunohistochemistry indicated that Fam20a protein was expressed in osteoid tissue and tooth germ of wild type mice (Figure 4A and C), but was not expressed in Fam20a–/– mice (P.0.d) (Figure 4B and D).
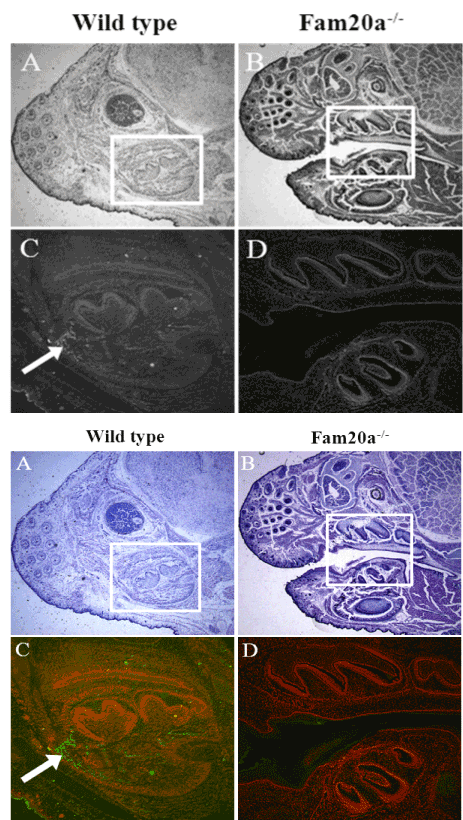
Figure 4. Immunohistochemistry.
Immunohistochemistry indicated that Fam20a protein was expressed in osteoid tissue and tooth germ of wild type mice, but was not expressed in Fam20a–/– mice (P.0.d).
Physiologic calcification or biomineralization of extracellular matrices is a normal developmental process that is essential for the proper formation and functioning of various tissues (eg, bones and teeth). If calcification or biomineralization process was disturbed, the formation of bone and tooth would also be disturbed and cause several diseases such as AI. To date, AI has been linked to mutations in six enamel proteins and proteinases (ENAM, AMBN, AMELX, MMP20, KLK4, and AMELOTIN), but many of the known AI kindreds do not have mutations in any of these genes [9,10]. In this study, we found typical AI and gum hypertrophy happened in Fam20a–/–mice. Recently, some studies have also reported that mutations of FAM20A gene were detected in AI patients [7,11]. Therefore we concluded that FAM20A played an important role in the regulation of biomineralization process.
Historically, use of the term AI was restricted to hereditary disorders characterized by abnormalities in the quantity and quality of enamel unaccompanied by metabolic or morphological defects in other body systems [12]. However, the definition of AI has been expanded to include heritable enamel defects that may be associated with lesions affecting other organ systems [12]. Additionally, some studies have reported that FAM20A Mutations Can Cause Enamel-Renal Syndrome [13,14]. So we will continue observing the systemic disorder of FAM20A KO mice in the future. Besides, we will also observe the histological change of FAM20A KO mice during different stage of tooth germ development to further clarify the function of FAM20A during the biomineralization process.
Taken together, the present study indicates that the loss of FAM20A in mice caused AI and gingival hyperplasia. This result is consisting with the previous report of AI and gingival hyperplasia caused by mutation of FAM20A genome in human beings. Further studies to clarify the relationship of AI and gingival hyperplasia with FAM20A in the mouse are next attempts we should confront.
- Witkop Jr CJ (1988) Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: problems in classification. J Oral Pathol 17: 547–553. [Crossref]
- Hu JC, Chun YH, Al Hazzazzi T, Simmer JP (2007) Enamel formation and amelogenesis imperfecta. Cells Tissues Organs (Print) 186: 78–85. [Crossref]
- Eastoe JE (1960) Organic matrix of tooth enamel. Nature 187: 411–412.
- Thesleff I (2003) Epithelial-mesenchymal signalling regulating tooth morphogenesis. J Cell Sci 116: 1647–1648. [Crossref]
- KimJW, Simmer JP, Lin BP, Seymen F, Bartlett JD,Hu JC (2006) Mutational analysis of candidate genes in 24 amelogenesis imperfecta families. Eur J Oral Sci 114(Suppl1): 3–12. [crossref]
- Nalbant D, Youn H, Nalbant SI, Sharma S, Cobos E, et al. (2005) FAM20: an evolutionarily conserved family of secreted proteins expressed in hematopoietic cells. BMC Genomics 6: 11.[Crossref]
- O’Sullivan J, Bitu CC, Daly SB, Urquhart JE, Barron MJ, et al. (2011) Whole-exome sequencing identifies FAM20A mutations as a cause of amelogenesis imperfecta and gingival hyperplasia syndrome. Am J Hum Genet 88: 616–620. [Crossref]
- An C, Ide Y, Nagano-Fujii M, Kitazawa S, Shoji I, et al. (2010) A transgenic mouse line with a 58-kb fragment deletion in chromosome 11E1 that encompasses part of the Fam20a gene and its upstream region shows growth disorder. Kobe J Med Sci 55: E82-92. [Crossref]
- Crawford PJ, Aldred M, Bloch-Zupan A (2007) Amelogenesis imperfecta. Orphanet J Rare Dis 2: 17. [Crossref]
- Santos MC, Hart PS, Ramaswami M, Kanno CM, Hart TC, et al. (2007) Exclusion of known gene for enamel development in two Brazilian families with amelogenesis imperfecta. Head Face Med 3: 8. [Crossref]
- Cho SH, Seymen F, Lee KE, Lee SK, Kweon YS, et al. (2012) Novel FAM20A mutations in hypoplastic amelogenesis imperfecta. Hum Mutat 33: 91-4. [Crossref]
- Aldred MJ, Savarirayan R, Crawford PJ (2003) Amelogenesis imperfecta: a classification and catalogue for the 21st century. Oral Dis 9: 19–23. [Crossref]
- Jaureguiberry G, De la Dure-Molla M, Parry D, Quentric M, Himmerkus N, et al. (2012) Nephrocalcinosis (enamel renal syndrome) caused by autosomal recessive FAM20A mutations. Nephron Physiol 122: 1-6. [Crossref]
- Wang SK, Aref P, Hu Y, Milkovich RN, Simmer JP, et al. (2013) FAM20A mutations can cause enamel-renal syndrome (ERS). PLoS Genet 9: e1003302. [Crossref]




