Abstract
Background: Norepinephrine (NE) dysregulation has been implicated as one of the mechanisms in the development of post-traumatic stress disorder (PTSD). Norepinephrine transporter (NET) plays an important role in regulating of NE level at the synapses. The gene of NET is a potential research target for PTSD. However, few studies have examined the genetic variants and DNA methylation of NET regards to the risk of PTSD.
Methods: We investigated 16 single-nucleotide polymorphisms (SNPs) across the NET whole gene in 506 veteran patients. Using a subset of individuals (161 patients) from our larger genetic dataset, promoter methylation was performed using pyrosequencing analysis.
Results: Significant difference was found in the genotype and allele frequencies for rs168924 and rs192303 by PTSD status, although the significance of rs192303 genotype (p=0.03) disappeared after Bonferroni correction. Neither rs168924 nor rs192303 was a significant predictor of PTSD diagnosis in primary logistic regression. However, significant interaction with Combat Exposure Scale (CES) was observed. The interaction effect was also positively associated with Clinician Administered PTSD Scale (CAPS). Higher CES and lower methylation were significantly associated with risk of PTSD, but no significant interaction effect was observed.
Conclusions: To our knowledge, this is the first study to extensively investigate NET gene Polymorphisms and promoter methylation among combat veteran PTSD. Our findings suggest the interaction of combat trauma and risk alleles of NET may account for the increased risk of PTSD. The NET promoter methylation may also participate in the process of PTSD development.
Key words
DNA methylation, epigenetics, genetic variants, norepinephrine transporter gene, polymorphisms, PTSD
Introduction
Post-traumatic stress disorder (PTSD) is a maladaptive response to life-threatening events, characterized by symptoms cluster of re-experiencing of the traumatic event, avoidance and numbing, negative alterations in cognitions and mood, and increased vigilance and arousal [1,2]. Trauma exposure is a required risk factor for development of PTSD, but is not, by itself, sufficient to cause PTSD, as not everyone exposed to trauma develops PTSD. Emerging research from various studies has indicated significant genetic contribution to individual susceptibility to PTSD [3]. However, the gene or genes participating in the PTSD process and the mode of inheritance remains undetermined.
Catecholamine norepinephrine (NE), also known as noradrenaline (NA), is the principal chemical messenger in central noradrenergic and peripheral sympathetic synapses, which plays a critical role in the mammalian response to stress [4]. Norepinephrine (NE) dysregulation has been implicated as the cause behind specific symptom clusters in the pathophysiology of PTSD [5,6]. However, the underlying molecular mechanism is not well understood. Approximately 80–90% of released norepinephrine is taken up again through the neuronal norepinephrine transporter (NET) [7,8]. The NET, also known as solute carrier family 6 member 2 (SLC6A2), present on the plasma membrane of noradrenergic neurons, limits the action of NE through reuptake into the cytoplasm [4]. The gene, with 14 exons, is located on human chromosome 16q12.2 [9]. NET belongs to the monoamine transporter superfamily and consists of 617 amino acids with 12 membrane-spanning domains [9,10]. The NET not only regulates the longevity of NE in the synapse but also plays an important role in presynaptic and postsynaptic homeostasis [11-13]. Given the fact that substantial evidence has suggested the existence of impaired neuronal norepinephrine reuptake in PTSD patients and the central role of NET in the regulation of central nervous system and peripheral norepinephrine turnover, NET should be a potential research target for PTSD.
Since noradrenergic neurotransmission can be regulated by changes in NET expression, it appears reasonable to suggest that NET genetic variations may confer risk for altered central NE system putatively contributing to affective disorder and /or treatment response [14]. A number of polymorphisms have been reported for the NET gene [15-17]. Some polymorphisms may lead to an altered transcriptional activity by changes in the DNA structure or function [16,18-20]. Some polymorphisms have been shown to have a functional effect on SLC6A2 gene expression [17]. Numerous single nucleotide polymorphisms (SNPS) within the SLC6A2 gene have been studied for association with psychiatric disorders, such as ADHD [15,20-23], major depression [24-26], panic disorder studies [27-29] and bipolar disorder [14], although there are mixed results. However, NET polymorphisms are poorly explored in PTSD. Only one letter report suggested that promoter SNP rs2242446 was independently associated with anxious arousal symptoms of PTSD and no positive association with the risk of PTSD was observed [30].
The complexity of the association between SLC6A2 and PTSD may be related, in part, to emerging evidence that not only genetic, but also epigenetic factors shape risk of mental illness. Epigenetic dysregulation has been implicated in pathogenesis of several psychiatric disorders such as depression [31], schizophrenia [32], eating disorders [33], and PTSD [34,35]. DNA methylation, an emerging epigenetic mechanism, may explain the potential mechanisms that accounts for how trauma exposure leads to sustained PTSD symptoms. The DNA methylation is often linked to regulation of gene expression – both transcriptional silencing and activation [35,36], and can be influenced by environmental factors [37,38]. The NET gene promoter region is extraordinarily rich in CpG islands and so is a primer target for DNA methylation [39]. There are no earlier reports regarding the association between NET promoter methylation and PTSD, although studies have reported findings in postural orthostatic tachycardia syndrome [39,40], major depression [41] and panic disorder [39].
To better elucidate the molecular basis shaping risk of PTSD at the SLC6A2 locus, we investigated whether variants within NET gene and promoter methylation are associated with the development of PTSD in veteran patients. We predicted that NET polymorphism and promoter methylation would differ by PTSD status and alleles’ differences and altered promote methylation of NET would be associated with PTSD symptom clusters.
Materials and methods
Participants
A total of 506 combat veterans were enrolled in the study, recruited through two Veterans Affairs Medical Centers (VAMC), the Cincinnati VAMC, Cincinnati, OH and Charleston VAMC, Charleston, SC (Table 1). Of the 506 participants in the study, 64.4% (n = 326) met criteria for PTSD. The majority of the participants were male (83.0%) and Caucasian (68.6%). The research protocol was approved by the Institutional Review Boards (IRB) of both the University of Cincinnati and Medical University of South Carolina. Subjects were recruited using similar methods at the two sites.
Procedure
Participants were assessed for the presence of PTSD and other major psychiatric diagnoses by a board-certified psychiatrist with either the Structured Clinical Interview for DSM-IV (SCID) [42] or the Mini-International Neuropsychiatric Interview (MINI) [43]. Full diagnostic level data and demographic data are available for all 506 participants. The Combat Exposure Scale (CES) is used to obtain information regarding exposure to wartime stressor events, with a higher number reflecting a higher severity of combat exposure. Participants were also interviewed using the Clinician Administered PTSD Scale (CAPS) [44] to assess symptoms’ frequency and intensity. All participants from the Charleston VAMC were recruited following this addition and a majority of participants from the Cincinnati VA in this present dataset. CAPS data is available for 404 of 506 participants and CES data is available for 478 out of 506 participants.
Inclusion/Exclusion criteria
Inclusion criteria included: 1) a history of combat exposure, as evidenced by a DD214 (i.e., official record of service) and 2) a report of combat exposure during the interview with a psychiatrist. In order to match the trauma-exposure severity between the PTSD subjects and controls, the majority of participants in the second half of this study were required to have CES score of 10 or above. Exclusion criteria included: current or lifetime DSM-IV schizophrenia, other psychotic disorders, bipolar disorder, and active substance abuse or dependence in the past six months. Combat veterans with comorbid major depression and anxiety disorders (as assessed during the SCID) were included. Individuals with a past history of substance abuse and dependence were also included if the last use of the substance was over 6 months prior to the enrollment.
SNP selection and genotyping
Genomic DNA was extracted from peripheral blood using a Wizard Genomic DNA purification kit (Promega, Madison, Wisconsin) following the manufacturer's protocol. 16 SNPs was randomly selected with minor allele frequencies of more than 0.1 to cover a region of 60 kb in the NET gene using a software tool (SNPbrowser version 4.0; Applied Biosystems) and a review of the literature with two exceptions: rs1805065 (MAF=0.0006) and rs5564 (MAF=0.07) since these two SNPs are located in special regions and has been studied in other disease (Bayles et al., 2012). The average spacing between SNPs across the gene was 3.7 kilobases (kb). The SNPs were genotyped by the TaqMan, probe-based SNP genotyping assay on the ABI StepOnePlus™ Real-Time PCR System ((Applied Biosystems, Foster City, CA).
Sodium Bisulfite Conversion and Pyrosequencing Analysis
Sodium bisulfite conversion of genomic DNA (20 μg) was obtained using Epitect Bisulphite kit (Qiagen), following the manufacturer's instructions. A total of 10 ng of modified DNA was subjected to PCR amplification of the specific promoter. Specially, two promoter regions and total 9 CpG sites across SLC6A2 CpG island were selected to do the PCR application and Pyrosequencing: Hs_SLC6A2_01_PM (ABI cat#: PM00173922) and Hs_SLC6A2_03_PM (ABI cat#: PM00173936). The primer sequences, the genomic location of the bisulfite pyrosequencing assays and the number of CpG sites investigated in each assay are shown in supplement 1. Following amplification, the biotinylated PCR products were purified and incubated with the sequencing primer designed to bind adjacent to the CpG sites of interest. Pyrosequencing was conducted using a PyroMark Q24 instrument (Qiagen), with subsequent quantitation of methylation levels determined with the PyroMark Q24 advance 3.0.0 software. Relative peak height differences were used to automatically calculate the percentage of methylated cytosines at each given site. The percent methylation fraction, i.e., the C/T ratio, was displayed above each CpG site in the analyzed sequence. Non-CpG cytosine residues were used as internal controls to verify efficient sodium bisulfite DNA conversion.
Statistical analysis
Statistical analysis was performed with SPSS software 21.0 (SPSS Inc. USA). Data distributions of all study variables were assessed for normality using the Shapiro-Wilk tests. Demographic and clinical variables were compared using chi-square χ2 test for categorical variables and nonparametric Independent Mann-Whitney U Test for continuous variables (age, CES, CAPS). Allelic and genotypic associations were analyzed using chi-square and chi-square linear-by-linear association test respectively. Main and interaction model of logistic regression analysis, adjusting for demographics, was used to assess the effect of SLC6A2 polymorphisms and methylation on the risk of PTSD. Main and interaction model of linear regression was also conducted to predict CAPS scores, controlling for demographics. The total methylation percentage across the 9 CpG sites was dichotomized based on the median value (median-split; 36.035) to improve the estimation stability of the logistic regression models [45]. Continuous variables including age, CAPS, and CES were centered to the mean. Given that we examined two related outcomes (one diagnostic and one quantitative phenotype) a Bonferroni correction was employed to adjust for multiple testing, and our p-value for determination of significance was p<0.025.
Haploview software (version 4.2; Broad Institute, Cambridge, Massachusetts, USA) was also used to analyze the status of pairwise linkage disequilibrium (LD) and haplotype frequencies.
Results
Demographic data and clinical features
A total of 506 subjects (180 combat controls, 326 PTSD) were enrolled in the study (table 1). The combat control and PTSD groups did not differ significantly in regards to race (χ2 [1, n = 506] = 2.288, p = 0.13) or gender χ2 [1, n = 506] = 2.509, p = 0.113. Average age was also not significantly different between combat control (M=43.22, SD=14.34) versus PTSD (M=43.07, SD=14.88), z=-0245, p=0.807). Of the PTSD cases, 211 (64.7%) patients also had a diagnosis of MDD. Due to study selection resulting in a quasi-complete separation (i.e., there were no cases of MDD in the control group), MDD was unable to be included as a covariate in analyses. Additionally, among participants with data for the CES (n=478), the level of combat trauma was significantly different between combat control (M=18.71, SD=8.70) versus PTSD (M=22.08, SD=8.21, Z=-3.95, p<0.001). Among participants with data for the CAPS (n=499), CAPS score was significantly different between combat control (M=11.67, SD=16.52) versus PTSD (M=78.13, SD=17.43), Z=-18.31, p<0.001).
Variable |
Combat Control (N= 180 ) |
PTSD (N=326 ) |
Age (years) |
M=43.22 |
M= 43.07 |
SD=14.34 |
SD= 14.88 |
Gender |
Male |
143 (79.7%) |
277(85.2 %) |
Female |
37 (20.3%) |
48 (14.8 %) |
Race |
EA |
133 (73.9%) |
218(66.9%) |
AA |
30 (16.7%) |
94 (28.8%) |
Others |
17 (9.4%) |
14 (4.3%) |
CES |
M=18.71 |
M=22.08 |
SD=8.70 |
SD=8.21 |
CAPS |
M=11.67 |
M=78.13 |
SD=16.52 |
SD=17.83 |
Major Depression, N (%) |
No Depresion |
180 (100%) |
115 (35.3%) |
Depression |
0 (0.00%) |
211 (64.7%) |
Table 1. Demographic and Clinical Characteristics. PTSD: posttraumatic stress disorder; CES: combat exposure scale; CAPS: clinician administered PTSD scale
Linkage disequilibrium and haplotype analysis
The LD map and block structure of the studied NET polymorphisms are presented in Figure 1. Five main haplotype blocks were detected from these 16 SNPs in our veteran population (Figure 1, Table 2). Block 1 was formed from rs1168924 and rs2242446 in the promoter region , which was in high LD (Figure 1). GT haplotype from rs168924 and rs2242446 was more frequent in PTSD than in control (p=0.0216) (Table 2). Block 2 was formed from rs192303 and rs36025 in the region of intron 1 to intron 2, which was in high LD (Figure 1). CC haplotype from rs192303 and rs36025 was more frequent in control than PTSD (p=0.0164) (Table 2). The results show that this region with intron 1 and intron 2 of the NET gene was associated with the development of PTSD. Block 3 was formed from the two loci rs36017 and rs5564 in the region of intron 3 to intron 5. Block 4 was formed rs47958, rs3785157, rs5569 and rs998424. Block 5 was formed rs1800887, rs15534 and rs7194256. Block 3, Block 4, and Block 5 were all in high LD (Figure 1), but no significant difference in haplotype frequencies were found between PTSD patients and combat controls (Table 2).
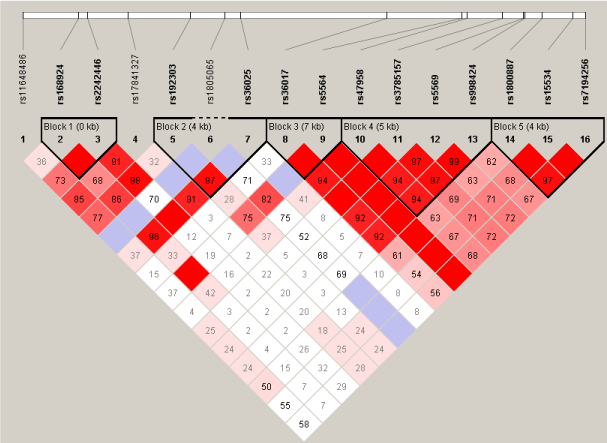
Figure 1. Linkage disequilibrium analyses: D' value of SNPs along the SLC6A2 gene, illustrating five haplotype blocks. D' value was calculated by Haploview version 4.2
Haplotype |
Frequency of haplotype |
|
Chi Square |
P Value |
|
Total |
Combat Control |
PTSD |
|
|
Block 1 |
AT |
0.569 |
0.595 |
0.554 |
1.603 |
0.2055 |
AC |
0.289 |
0.296 |
0.285 |
0.15 |
0.6982 |
GT |
0.142 |
0.108 |
0.161 |
5.28 |
0.0216 |
Block 2 |
CC |
0.712 |
0.758 |
0.687 |
5.756 |
0.0164 |
GC |
0.165 |
0.148 |
0.174 |
1.179 |
0.2775 |
GT |
0.121 |
0.094 |
0.136 |
3.794 |
0.0514 |
Block 3 |
GA |
0.553 |
0.569 |
0.543 |
0.645 |
0.4218 |
CA |
0.387 |
0.369 |
0.397 |
0.719 |
0.3966 |
CG |
0.061 |
0.061 |
0.06 |
0.003 |
0.9552 |
Block 4 |
ACGT |
0.417 |
0.401 |
0.427 |
0.655 |
0.4184 |
CCGT |
0.289 |
0.294 |
0.287 |
0.053 |
0.8184 |
CTAC |
0.269 |
0.289 |
0.258 |
1.096 |
0.2952 |
Block 5 |
TCC |
0.733 |
0.747 |
0.725 |
0.534 |
0.4648 |
CTT |
0.187 |
0.167 |
0.198 |
1.485 |
0.223 |
CCC |
0.076 |
0.081 |
0.074 |
0.183 |
0.6687 |
Table 2. Haplotype analysis of SLC6A2 gene in combat control and PTSD. PTSD: posttraumatic stress disorder; SLC6A2: solute carrier family 6 member 2
Allele and Genotype frequency of the SLC6A2 polymorphisms
To assess the influence of each NET variant on the incidence of PTSD, we performed further analyses. rs1800887 has significant deviation from Hardy-Weinberg equilibrium obverted in the populations and were not used in the later analysis (Supplement .2). Two SNPs (rs168924, rs192303) showed nominally significant allelic association with PTSD (χ2 =5.28, p=0.0216; χ2 =5.28, p=0.0216,) (Table 3). A significant association was found between the genotype frequency of rs168924 and PTSD (χ2 =5.244, p=0.022,) and weak genotypic association for rs192303 was also observed (χ2 =4.724, p=0.03, not significant after Bonferroni correction) (Table 3). No statistically significant differences either in the allele frequencies or in genotype frequencies for other NET polymorphisms were detected. When selecting the subset of participants who were Caucasian (n=351), only rs192303 has weak difference in allele and genotypic frequency by PTSD status (p=0.0325, p=0.027 respectively).
| |
Genotype Count (Frequency) |
Allele Count( frequency) |
| |
|
Non PTSD |
PTSD |
|
|
|
Non PTSD |
PTSD |
|
|
| |
|
n |
% |
n |
% |
χ2 |
p |
|
n |
% |
n |
% |
χ2 |
p |
rs116484836 |
CC |
127 |
70.6% |
218 |
66.9% |
|
|
|
|
|
|
|
|
|
TC |
47 |
26.1% |
98 |
30.1% |
0.465 |
0.495 |
C |
301 |
83.6% |
534 |
81.9% |
0.47 |
0.4932 |
TT |
6 |
3.3% |
10 |
3.0% |
|
|
T |
59 |
16.4% |
118 |
18.1% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rsA168924 |
AA |
143 |
79.4% |
232 |
71.2% |
|
|
|
|
|
|
|
|
|
AG |
35 |
19.4% |
82 |
25.1% |
5.244 |
0.022 |
A |
321 |
89.2% |
547 |
83.9% |
5.28 |
0.0216 |
GG |
2 |
1.1% |
12 |
3.7% |
|
|
G |
39 |
10.8% |
105 |
16.1% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs2242446 |
CC |
13 |
7.3% |
28 |
8.6% |
|
|
|
|
|
|
|
|
|
TC |
80 |
44.7% |
129 |
39.7% |
0.149 |
0.699 |
C |
106 |
29.6% |
185 |
28.5% |
0.148 |
0.7004 |
TT |
86 |
48.0% |
168 |
51.7% |
|
|
T |
252 |
70.4% |
465 |
71.5% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs17841327 |
CC |
61 |
34.0% |
108 |
33.2% |
|
|
|
|
|
|
|
|
|
AC |
93 |
52.0% |
168 |
51.7% |
0.099 |
0.753 |
C |
215 |
60.1% |
384 |
59.1% |
|
|
AA |
25 |
14.0% |
49 |
15.1% |
|
|
A |
143 |
39.9% |
266 |
40.9% |
0.092 |
0.762 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs192303 |
CC |
104 |
57.8% |
155 |
47.5% |
|
|
|
|
|
|
|
|
|
GC |
65 |
36.1% |
140 |
42.9% |
4.724 |
0.03 |
C |
273 |
75.8% |
450 |
69.0% |
5.28 |
0.0216 |
GG |
11 |
6.1% |
31 |
9.6% |
|
|
G |
87 |
24.2% |
202 |
31.0% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs1805065 |
TT |
0 |
0.0% |
0 |
0.0% |
|
|
|
|
|
|
|
|
|
CT |
6 |
3.3% |
7 |
2.1% |
0.650 |
0.420 |
T |
6 |
1.7% |
7 |
1.1% |
0.643 |
0.4225 |
TT |
180 |
96.7% |
326 |
97.9% |
|
|
C |
354 |
98.3% |
645 |
98.9% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs36025 |
CC |
148 |
82.2% |
245 |
75.4% |
|
|
|
|
|
|
|
|
|
TC |
30 |
16.7% |
70 |
21.5% |
3.849 |
0.050 |
C |
326 |
90.6% |
560 |
86.1% |
4.168 |
0.0412 |
TT |
2 |
1.1% |
10 |
3.1% |
|
|
T |
34 |
9.4% |
90 |
13.9% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs36017 |
GG |
59 |
32.8% |
94 |
28.9% |
|
|
|
|
|
|
|
|
|
CG |
87 |
48.3% |
165 |
50.8% |
0.656 |
0.418 |
G |
205 |
56.9% |
353 |
54.3% |
0.651 |
0.4196 |
CC |
34 |
18.9% |
66 |
20.3% |
|
|
C |
155 |
43.1% |
297 |
45.7% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs5564 |
GG |
1 |
0.6% |
4 |
1.2% |
|
|
|
|
|
|
|
|
|
AG |
20 |
11.1% |
31 |
9.6% |
0.003 |
0.955 |
G |
22 |
6.1% |
39 |
6.0% |
0.003 |
0.9529 |
AA |
159 |
88.3% |
289 |
89.2% |
|
|
A |
338 |
93.9% |
609 |
94.0% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs47958 |
CC |
64 |
35.6% |
104 |
31.9% |
|
|
|
|
|
|
|
|
|
AC |
86 |
47.8% |
161 |
49.4% |
0.769 |
0.380 |
C |
214 |
59.4% |
369 |
56.6% |
0.771 |
0.3799 |
AA |
30 |
16.6% |
61 |
18.7% |
|
|
A |
146 |
40.6% |
283 |
43.4% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs3785157 |
TT |
15 |
8.3% |
28 |
8.7% |
|
|
|
|
|
|
|
|
|
CT |
76 |
42.2% |
113 |
35.1% |
1.128 |
0.288 |
T |
106 |
29.4% |
169 |
26.2% |
1.191 |
0.2752 |
CC |
89 |
49.5% |
181 |
56.2% |
|
|
C |
254 |
70.6% |
475 |
73.8% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs5569 |
AA |
15 |
8.4% |
31 |
9.6% |
|
|
|
|
|
|
|
|
|
GA |
77 |
43.0% |
119 |
37.0% |
0.341 |
0.559 |
A |
107 |
29.9% |
181 |
28.1% |
0.357 |
0.5502 |
GG |
87 |
48.6% |
172 |
53.4% |
|
|
G |
251 |
70.1% |
463 |
71.9% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs998424 |
CC |
16 |
8.9% |
31 |
9.5% |
|
|
|
|
|
|
|
|
|
TC |
77 |
42.8% |
1120 |
37.0% |
0.58 |
0.455 |
C |
109 |
30.3% |
182 |
28.0% |
0.586 |
0.4439 |
TT |
87 |
48.3% |
174 |
53.5% |
|
|
T |
251 |
69.7% |
468 |
72.0% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs15534 |
CC |
127 |
70.5% |
212 |
65.0% |
|
|
|
|
|
|
|
|
|
TC |
46 |
25.6% |
99 |
30.4% |
1.403 |
0.236 |
C |
300 |
83.3% |
523 |
82.2% |
1.485 |
0.223 |
TT |
7 |
3.9% |
15 |
4.6% |
|
|
T |
60 |
16.7% |
129 |
19.8% |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
rs7194256 |
CC |
126 |
70.4% |
210 |
64.8% |
|
|
|
|
|
|
|
|
|
TC |
44 |
24.6% |
98 |
30.2% |
1.033 |
0.310 |
C |
296 |
82.7% |
521 |
79.9% |
1.124 |
0.2891 |
TT |
9 |
5.0% |
16 |
5.0% |
|
|
T |
62 |
17.3% |
127 |
20.1% |
|
|
Table 3. Comparison of SLC6A2 Gene Genotype and Allele Frequencies among Combat Control and PTSD Individuals. PTSD: posttraumatic stress disorder; SLC6A2: solute carrier family 6 member 2
Multivariate logistic regression analysis for the influence of allele types of SLC6A2 and Combat Exposure Scale (CES) on the risk of PTSD diagnosis
Logistic regression analysis was performed to look at if CES and associated SNPs would affect the risk of PTSD after controlling demographics (4). SNP rs168924 and rs192303 was entered into the model separately. In the main effects model, CES was strongly associated with PTSD status (OR=1.052, 95% CI=1.027-1.078, p<0.001), but allele types of rs168924 (p=0.090) and rs192303 (p=0.094) were not significant predictor of PTSD diagnosis after adjusting for age, sex and race. In the interaction model, the interaction effect of SNPxCES was significant (OR=1.075, 95% CI=1.012-1.142, p=0.019; OR=1.056, 95% CI=1.014-1.099, p=0.009 respectively). The interaction model also provided a significantly improved fit over the main effects model (χ2 (1) =6.213, p=0.013, χ2 (1) =7.273, p=0.007, respectively). So persons with more traumatic events and with more risk alleles of rs168924 or rs192303 might be at increased risk for PTSD.
Multivariable linear regression analysis for the association of allele types of SLC6A2 and Combat Exposure Scale (CES) on the PTSD symptom severity
Multivariable linear regression analysis was also conducted to control other variables (table 5). In the main model, rs168924, rs192303 allele types were positively associated with CAPS score (β=0.1, p=0.029, β=0.085, p=0.062), but not statistically significant. In the interaction model, the interaction term for SNP X CES was also positively associated with CAPS Score but rs192303xCES not significant after Bonferroni correction (β=0.114, p=0.024; β=0.128, p=0.034, respectively). Based on the adjusted R2, both of the interaction models also explained 7% more of the variance in PTSD symptom severity than the main effects models. CES was always a significant predictor of PTSD symptom severity (p<0.0001). So persons with more traumatic events and with more risk alleles of rs168924 or rs192303 may develop more severe symptoms in PTSD.
|
Main effects model |
|
Interaction model |
|
OR |
95% CI of OR |
p |
|
OR |
95% CI of OR |
p |
Age |
0.998 |
0.985 |
1.011 |
0.778 |
|
1 |
0.987 |
1.013 |
0.991 |
Gender |
0.913 |
0.552 |
1.512 |
0.724 |
|
0.917 |
0.553 |
1.522 |
0.738 |
Race |
1.319 |
0.849 |
2.049 |
0.218 |
|
1.304 |
0.836 |
2.032 |
0.242 |
CES |
1.052 |
1.027 |
1.078 |
0 |
|
1.037 |
1.01 |
1.065 |
0.006 |
rs168924 |
1.425 |
0.946 |
2.147 |
0.09 |
|
1.575 |
1.005 |
2.467 |
0.047 |
rs168924 X CES |
|
|
|
|
|
1.075 |
1.012 |
1.142 |
0.019 |
Age |
0.999 |
0.985 |
1.012 |
0.828 |
|
0.999 |
0.986 |
1.013 |
0.929 |
Gender |
0.899 |
0.542 |
1.49 |
0.679 |
|
0.882 |
0.531 |
1.465 |
0.628 |
Race |
1.348 |
0.871 |
2.084 |
0.18 |
|
1.382 |
0.89 |
2.147 |
0.15 |
CES |
1.052 |
1.027 |
1.078 |
0 |
|
1.024 |
0.993 |
1.056 |
0.125 |
rs192303 |
1.303 |
0.956 |
1.776 |
0.094 |
|
1.412 |
1.02 |
1.954 |
0.038 |
rs192303 X CES |
|
|
|
|
|
1.056 |
1.014 |
1.099 |
0.009 |
Table 4. Effect of SLC6A2 SNP genotype and Combat Severity Score (CES)on risk of lifetime PTSD diagnosis. PTSD: posttraumatic stress disorder; SLC6A2: solute carrier family 6 member 2; SNP: single nucleotide polymorphism. Continuous variables were centered to the mean.
|
Main effects model |
Interaction model |
|
b |
beta |
t |
p |
b |
beta |
t |
p |
Age |
0.017 |
0.007 |
0.148 |
0.882 |
0.034 |
0.013 |
0.299 |
0.765 |
Gender |
-4.215 |
-0.043 |
-0.953 |
0.341 |
-4.373 |
-0.045 |
-0.992 |
0.321 |
Race |
1.122 |
0.018 |
0.4 |
0.689 |
1.14 |
0.019 |
0.408 |
0.683 |
CES |
0.926 |
0.214 |
4.687 |
<0.0001 |
0.701 |
0.162 |
3.179 |
0.002 |
rs168924 |
7.221 |
0.1 |
2.193 |
0.029 |
6.227 |
0.087 |
1.883 |
0.06 |
rs168924 X CES |
|
|
|
|
0.902 |
0.114 |
2.259 |
0.024 |
Age |
0.023 |
0.009 |
0.205 |
0.838 |
0.025 |
0.01 |
0.221 |
0.825 |
Gender |
-4.635 |
-0.048 |
-1.047 |
0.296 |
-4.956 |
-0.051 |
-1.123 |
0.262 |
Race |
1.559 |
0.025 |
0.56 |
0.576 |
2.192 |
0.036 |
0.785 |
0.433 |
CES |
0.927 |
0.214 |
4.684 |
<0.0001 |
0.554 |
0.128 |
2.101 |
0.036 |
rs192303 |
4.861 |
0.085 |
1.873 |
0.062 |
4.776 |
0.084 |
1.846 |
0.065 |
rs192303 X CES |
|
|
|
|
0.654 |
0.128 |
2.121 |
0.034 |
Table 5. Effect of SLC6A2 SNP genotype and Combat Severity Score (CES) on PTSD symptom severity. PTSD: posttraumatic stress disorder; SLC6A2: solute carrier family 6 member 2; SNP: single nucleotide polymorphism. Continuous variables were centered to the mean.
Effect of SLC6A2 methylation and Combat Exposure Scale (CES) on the risk of PTSD diagnosis
A total of 161 samples (57 combat controls, 104 PTSD) were used to do the DNA methylation sequencing analysis. These patients were all recently recruited from Charleston VAMC and relatively young.Multivariate logistic regression analysis was performed for the influence of SLC6A2 methylation and CES on the risk of PTSD diagnosis (Table 6). In the main model, Methylation group is a significant predictor of risk of PTSD (OR=0.383, 95% CI=0.171-0.857, p=0.020), indicating that low methylation showed higher risk of PTSD after controlling for demographics. Although higher CES also showed increased risk of PTSD (OR=1.077, 95% CI=1.031-1.125, p=0.001), the interaction model, the interaction term for Methylation X CES was not significant. So persons with low methylation level may independently have higher risk of PTSD.
|
|
OR |
95% CI of OR |
p |
Age |
|
1.009 |
0.954 |
1.068 |
0.752 |
Gender |
|
0.793 |
0.113 |
5.58 |
0.816 |
Race |
|
2.29 |
1.083 |
4.84 |
0.03 |
CES |
|
1.077 |
1.031 |
1.125 |
0.001 |
Methylation Groupa |
0.383 |
0.171 |
0.857 |
0.02 |
Table 6. Effect of SLC6A2 Methylation and Combat Severity Score (CES) on risk of lifetime PTSD diagnosis. PTSD: posttraumatic stress disorder; SLC6A2: solute carrier family 6 member. Continuous variables were centered to the mean. aMedian-Split
Discussion
To our knowledge, this is the first report of SNP studies spanning the full spectrum of SLC6A2 gene and also the first report of SLC6A2 promoter methylation studies in peripheral blood of veterans with and without PTSD. We tested a total of 16 SNPS and found that promoter SNP rs168924 and intron 1 SNP rs192303 were linked to the susceptibility to PTSD.Our findings indicate that veterans with more traumatic events and with more risk alleles of rs168924 or rs192303 may be at increased risk for PTSD development. Veterans with low methylation level in SLC6A2 promoter region may independently have higher risk of PTSD.To argue against false positive findings, we analyzed the data with different statistic approaches. Our positive findings hold true regardless of analyzing approaches.
The 5′-flanking promoter region of the NET gene contains several cis-elements that play a critical role in transcription regulation [46,47], so changes in this promoter DNA structure may lead to an altered transcriptional activity responsible for predisposition to PTSD. Therefore, the polymorphisms in this region are very important candidates for genetic studies in PTSD. Three promoter SNPs rs11648486, rs168924 and rs2242446 were detected in our studies. Only rs168924 was found to be significantly associated with risk of PTSD. Although we failed to detect a significant association between rs2242446 and risk of PTSD, we found the rs168924 is in LD with rs2242446, which consistent with other studies [28]. GT haplotype from rs1168924 and rs2242446 was significantly more frequent in PTSD than in controls. The results show that this promoter region of the NET gene was associated with the development of PTSD. SNP rs192303 is located in intron 1, a region reported to be responsible for high-level transcription of NET [46]. In combination with the promoter region, the intron 1 region contributed to the highest level of transcriptional activity within noradrenergic cells [47]. So it is biologically plausible that this SNP or variant that is in high LD with this SNP may affect NET expression and thus increase the risk of developing PTSD.
Since coding polymorphisms in the NET gene are rare, most did not pass the selection criteria as they were less polymorphic (MAF >=0.1) and were therefor not included in our studies. One SNP (rs1805065), located in exon 2 was selected in our studies according to other research findings [40]. Another SNP, rs5564, located in the spice site consensus sequence of exon 5 were included, because T allele was found to be significantly associated with the diagnosis of Postural Orthostatic Tachycardia Syndrome (POTs) [40]. However, we did not find significant association of both SNPs with PTSD.
A silent rs5569 polymorphism, located at exon 9 of the NET gene, was a particularly interesting candidate because it has higher heterozygosity than the other markers [19,29]. The G/G genotype was found to be correlated with the concentration of 3-methoxy-4-hydroxyphenylglycol (MHPG), a primary NE metabolite in the cerebrospinal fluid, consequently attributing to the increased reuptake of NE [48]. So we also first time studies this SNP in PTSD. However, no apparent relations supposed were observed between this polymorphism and PTSD. We found that rs3785157, rs5569, rs998424 are in high LD which was in line with other studies [22]. But the results for these three SNPs were not significant in our studies.
We also determine the differences in NET gene promoter methylation between traumas exposed individuals with and without PTSD for the first time. We found that lower NET promoter methylation may be a risk for the development of PTSD. Our study is consistent to other studies, which reported hypo-methylation in depressed and panic disorder patients [39,41]. However, we didn’t find that SLC6A2 promoter methylation was correlated with PTSD symptom severity scales. We didn’t find that combat trauma had influence on DNA methylation neither and there’s no interaction effect of CES and NET promoter methylation on risk of PTSD in our studies. The region around the transcription start site is transcriptionally important, and contains multiple putative transcription factor binding sites [40,49,50]. The analyzed region we chose is Chr16: 55690108 to Chr16: 55690516, which encompasses transcription start site (TSS) chr16:55690555. It is possible that this methylation island could have impact on gene expression as the methylation site is just located at the promoter region.
Our results indicate that considering both genetic variants and DNA methylation will improve the explanation of PTSD even though we didn’t find the direct interaction effect of these two factors in our sample size. Genetic variant and DNA methylation may jointly contribute to gene expression or alternative splicing [51].
Limitations
First, the selected16 SNPs markers may not provide complete coverage of the NET gene because the r2 value between some contiguous markers was less than 0.8 [52]. In addition, our random SNP selection method may decrease the power of the study compared with tag SNP selection method [53]. Second, we used the whole blood to carry out the methylation studies in PTSD. Despite DNA methylation patterns between different people, and between different tissues within an individual were found to be highly conserved [54], it cannot be ruled out that SLC6A2 DNA methylation may differ specifically in diseased central neurons. Third, our sample size is relatively small. The positive findings have to be considered explorative and findings need to be further verified in larger samples.
Conclusion
This is the first study to determine the role of both genetic and epigenetic factors within the genomic region of the SLC6A2 gene on the risk of PTSD. Our results suggest that genetic variants and promoter methylation in the SLC6A2 might be associated with the susceptibility of PTSD. Further in-depth analysis of SNPs and DNA methylation on transcriptional regulation of the SLC6A2 gene would be of value.
Acknowledgements
This research was supported by the Ralph H. Johnson VA Medical Center and the Clinical Sciences Program of the Department of Veterans Affairs (Merit Review Grant No.1I01CX000487-01A1)
The views expressed in this article are those of the author(s) and do not necessarily represent the views of the Department of Veterans Affairs. All research performed was in compliance with the code of ethics of the World Medical Association (Declaration of Helsinki) and the standards established by the Ralph H. Johnson VA Medical Center Institutional Review Road. We gratefully acknowledge the assistance of Joshua Voltin for his contribution in the proof-reading and data verification.
Conflict of interest
The authors have no conflicts of interest to disclosure.
References
- Rabe-Jablonska J, B2021 Copyright OAT. All rights reserv in the fourth edition of the classification of mental disorders prepared by the American Psychiatric Association: diagnostic and statistical manual of mental disorders (DMS-IV -- options book]. Psychiatr Pol 28: 255-268.
- Segman RH, Shefi N, Goltser-Dubner T, Friedman N, Kaminski N, et al. (2005) Peripheral blood mononuclear cell gene expression profiles identify emergent post-traumatic stress disorder among trauma survivors. Mol Psychiatry 10: 500-513. [Crossref]
- True WR, Rice J, Eisen SA, Heath AC, Goldberg J, et al. (1993) A twin study of genetic and environmental contributions to liability for posttraumatic stress symptoms. Arch Gen Psychiatry 50: 257-264. [Crossref]
- Mandela P, Ordway GA (2006) The norepinephrine transporter and its regulation. J Neurochem 97: 310-333. [Crossref]
- O'Donnell T, Hegadoren KM, Coupland NC (2004) Noradrenergic mechanisms in the pathophysiology of post-traumatic stress disorder. Neuropsychobiology 50: 273-283. [Crossref]
- Strawn JR, Geracioti TD Jr (2008) Noradrenergic dysfunction and the psychopharmacology of posttraumatic stress disorder. Depress Anxiety 25: 260-271. [Crossref]
- Esler M, Jennings G, Lambert G, Meredith I, Horne M, et al. (1990) Overflow of catecholamine neurotransmitters to the circulation: source, fate, and functions. Physiol Rev 70: 963-985. [Crossref]
- Schroeder C, Jordan J (2012) Norepinephrine transporter function and human cardiovascular disease. Am J Physiol Heart Circ Physiol 303: H1273-1282. [Crossref]
- Brüss M, Kunz J, Lingen B, Bönisch H (1993) Chromosomal mapping of the human gene for the tricyclic antidepressant-sensitive noradrenaline transporter. Hum Genet 91: 278-280. [Crossref]
- Bonisch H, Runkel F, Roubert C, Giros B, Bruss M (1999) The human desipramine-sensitive noradrenaline transporter and the importance of defined amino acids for its function. J Auton Pharmacol 19: 327-333. [Crossref]
- Axelrod J, Kopin IJ (1969) The uptake, storage, release and metabolism of noradrenaline in sympathetic nerves. Prog Brain Res 31: 21-32. [Crossref]
- Iversen LL (1971) Role of transmitter uptake mechanisms in synaptic neurotransmission. Br J Pharmacol 41: 571-591. [Crossref]
- Zahniser NR, Doolen S (2001) Chronic and acute regulation of Na+/Cl- -dependent neurotransmitter transporters: drugs, substrates, presynaptic receptors, and signaling systems. Pharmacol Ther 92: 21-55.
- Chang CC, Lu RB, Ma KH, Chang HA, Chen CL, et al. (2007) Association study of the norepinephrine transporter gene polymorphisms and bipolar disorder in Han Chinese population. World J Biol Psychiatry 8: 188-195. [Crossref]
- Dlugos AM, Hamidovic A, Palmer AA, de Wit H (2009) Further evidence of association between amphetamine response and SLC6A2 gene variants. Psychopharmacology (Berl) 206: 501-511. [Crossref]
- Kim CH, Hahn MK, Joung Y, Anderson SL, Steele AH, et al. (2006) A polymorphism in the norepinephrine transporter gene alters promoter activity and is associated with attention-deficit hyperactivity disorder. Proc Natl Acad Sci U S A 103: 19164-19169. [Crossref]
- Hahn MK, Mazei-Robison MS, Blakely RD (2005) Single nucleotide polymorphisms in the human norepinephrine transporter gene affect expression, trafficking, antidepressant interaction, and protein kinase C regulation. Mol Pharmacol 68: 457-466. [Crossref]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, et al. (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet 22: 239-247. [Crossref]
- Stober G, Nothen MM, Porzgen P, Bruss M, Bonisch H, et al. (1996) Systematic search for variation in the human norepinephrine transporter gene: identification of five naturally occurring missense mutations and study of association with major psychiatric disorders. Am J Med Genet 67: 523-532. [Crossref]
- Zill P, Engel R, Baghai TC, Juckel G, Frodl T, et al. (2002) Identification of a naturally occurring polymorphism in the promoter region of the norepinephrine transporter and analysis in major depression. Neuropsychopharmacology 26: 489-493. [Crossref]
- Hahn MK, Steele A, Couch RS, Stein MA, Krueger JJ (2009) Novel and functional norepinephrine transporter protein variants identified in attention-deficit hyperactivity disorder. Neuropharmacology 57: 694-701. [Crossref]
- Kim JW, Biederman J, McGrath CL, Doyle AE, Mick E, et al. (2008) Further evidence of association between two NET single-nucleotide polymorphisms with ADHD. Mol Psychiatry 13: 624-630. [Crossref]
- Thakur GA, Sengupta SM, Grizenko N, Choudhry Z, Joober R (2012) Comprehensive phenotype/genotype analyses of the norepinephrine transporter gene (SLC6A2) in ADHD: relation to maternal smoking during pregnancy. PLoS One 7: e49616. [Crossref]
- Chang CC, Lu RB, Chen CL, Chu CM, Chang HA, et al. (2007) Lack of association between the norepinephrine transporter gene and major depression in a Han Chinese population. J Psychiatry Neurosci 32: 121-128. [Crossref]
- Kim YK, Hwang JA1, Lee HJ1, Yoon HK1, Ko YH1, et al. (2014) Association between norepinephrine transporter gene (SLC6A2) polymorphisms and suicide in patients with major depressive disorder. J Affect Disord 158: 127-132. [Crossref]
- Zhao X, Huang Y, Ma H, Jin Q, Wang Y, et al. (2013) Association between major depressive disorder and the norepinephrine transporter polymorphisms T-182C and G1287A: a meta-analysis. J Affect Disord 150: 23-28. [Crossref]
- Buttenschøn HN, Kristensen AS, Buch HN, Andersen JH, Bonde JP, et al. (2011) The norepinephrine transporter gene is a candidate gene for panic disorder. J Neural Transm (Vienna) 118: 969-976. [Crossref]
- Lee YJ, Hohoff C, Domschke K, Sand P, Kuhlenbäumer G, et al. (2005) Norepinephrine transporter (NET) promoter and 5'-UTR polymorphisms: association analysis in panic disorder. Neurosci Lett 377: 40-43. [Crossref]
- Sand PG, Mori T, Godau C, Stöber G, Flachenecker P, et al. (2002) Norepinephrine transporter gene (NET) variants in patients with panic disorder. Neurosci Lett 333: 41-44. [Crossref]
- Pietrzak RH, Sumner JA, Aiello AE, Uddin M, Neumeister A, et al. (2015) Association of the rs2242446 polymorphism in the norepinephrine transporter gene SLC6A2 and anxious arousal symptoms of posttraumatic stress disorder. J Clin Psychiatry 76: e537-538. [Crossref]
- Hillemacher T, Frieling H, Muschler MA, Bleich S (2007) Homocysteine and epigenetic DNA methylation: a biological model for depression? Am J Psychiatry 164: 1610. [Crossref]
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, et al. (2006) Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet 15: 3132-3145. [Crossref]
- Frieling H, Gozner A, Römer KD, Lenz B, Bönsch D, et al. (2007) Global DNA hypomethylation and DNA hypermethylation of the alpha synuclein promoter in females with anorexia nervosa. Mol Psychiatry 12: 229-230. [Crossref]
- Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, et al. (2011) Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet 156B: 700-708. [Crossref]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, et al. (2010) Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci U S A 107: 9470-9475. [Crossref]
- Zieker J, Zieker D, Jatzko A, Dietzsch J, Nieselt K, et al. (2007) Differential gene expression in peripheral blood of patients suffering from post-traumatic stress disorder. Mol Psychiatry 12: 116-118. [Crossref]
- Aguilera O, Fernández AF, Muñoz A, Fraga MF (2010) Epigenetics and environment: a complex relationship. J Appl Physiol (1985) 109: 243-251. [Crossref]
- Jaenisch R, Bird A (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33 Suppl: 245-254. [Crossref]
- Esler M, Alvarenga M, Pier C, Richards J, El-Osta A, et al. (2006) The neuronal noradrenaline transporter, anxiety and cardiovascular disease. J Psychopharmacol 20: 60-66. [Crossref]
- Bayles R, Harikrishnan KN, Lambert E, Baker EK, Agrotis A, et al. (2012) Epigenetic modification of the norepinephrine transporter gene in postural tachycardia syndrome. Arterioscler Thromb Vasc Biol 32: 1910-1916. [Crossref]
- Bayles R, Baker EK, Jowett JB, Barton D, Esler M, et al. (2013) Methylation of the SLC6a2 gene promoter in major depression and panic disorder. PLoS One 8: e83223. [Crossref]
- First MB (1997) User's guide for the structured clinical interview for DSM-IV axis II personality disorders : SCID-II. Washington, DC: American Psychiatric Press.
- Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, et al. (1998) The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry 59: 22-33. [Crossref]
- Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, et al. (1995) The development of a Clinician-Administered PTSD Scale. J Trauma Stress 8: 75-90. [Crossref]
- Chang SC, Koenen KC, Galea S, Aiello AE, Soliven R, et al. (2012) Molecular variation at the SLC6A3 locus predicts lifetime risk of PTSD in the Detroit Neighborhood Health Study. PLoS One 7: e39184. [Crossref]
- Kim CH, Kim HS, Cubells JF, Kim KS (1999) A previously undescribed intron and extensive 5' upstream sequence, but not Phox2a-mediated transactivation, are necessary for high level cell type-specific expression of the human norepinephrine transporter gene. J Biol Chem 274: 6507-6518. [Crossref]
- Meyer J, Wiedemann P, Okladnova O, Brüss M, Staab T, et al. (1998) Cloning and functional characterization of the human norepinephrine transporter gene promoter. J Neural Transm (Vienna) 105: 1341-1350. [Crossref]
- Jonsson EG, Nothen MM, Gustavsson JP, Neidt H, Bunzel R, et al. (1998) Polymorphisms in the dopamine, serotonin, and norepinephrine transporter genes and their relationships to monoamine metabolite concentrations in CSF of healthy volunteers. Psychiatry Res 79: 1-9. [Crossref]
- Bayles R, Baker E, Eikelis N, El-Osta A, Lambert G (2010) Histone modifications regulate the norepinephrine transporter gene. Cell Cycle 9: 4600-4601. [Crossref]
- Harikrishnan KN, Bayles R, Ciccotosto GD, Maxwell S, Cappai R, et al. (2010) Alleviating transcriptional inhibition of the norepinephrine slc6a2 transporter gene in depolarized neurons. J Neurosci 30: 1494-1501. [Crossref]
- Soto-Ramírez N, Arshad SH, Holloway JW, Zhang H, Schauberger E, et al. (2013) The interaction of genetic variants and DNA methylation of the interleukin-4 receptor gene increase the risk of asthma at age 18 years. Clin Epigenetics 5: 1. [Crossref]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. (2002) The structure of haplotype blocks in the human genome. Science 296: 2225-2229. [Crossref]
- Stram DO (2004) Tag SNP selection for association studies. Genet Epidemiol 27: 365-374. [Crossref]
- Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, et al. (2009) Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet 18: 4808-4817. [Crossref]

