Keywords
ebola disease virus, West-Africa epidemic, mobile laboratory deployment
Deployment of IPD laboratory in Guinea
The unit of Arbovirus and Haemorrhagic Fever Viruses at the Institut Pasteur de Dakar (IPD), a WHO-approved collaborating Centre was the first laboratory deployed to Conakry in the Ebola virus disease (EVD) outbreak in West-Africa. On 20 March 2014, the IPD laboratory received a letter from the WHO and the Guinean Ministry of Health, informing about a suspected haemorrhagic fever outbreak and difficulties to send collected samples to the IPD. They therefore requested the deployment of experts to Guinea for technical support in order to diagnose the haemorrhagic fever of unknown origin. The outbreak was identified by the Institut Pasteur (France) on 21 March 2014 [1,2] in samples shipped to France by a Médecins sans Frontières investigation team.
On 23 March 2014, an IPD team was deployed in Conakry, at Donka Hospital, and was the first laboratory arriving in Guinea. The same day, they analysed samples from Macenta, Guéckédou, Kissidougou and confirmed EBOV as responsible for the outbreak. The IPD laboratory detected the first EVD cases in Conakry on 26 March and in Liberia on 29 March 2014.
Laboratory structure and procedures
The Institut Pasteur de Dakar mobile laboratory (IPD-ML) was set up in Donka hospital in the Infectious Disease Department. The IPD ML operated daily 14 hours from 7.00 to 21.00 and sometimes until late at night. It was organized in pre-analytical, analytical and post-analytical areas.
For the pre-analytical step, a reception area for samples collected at Ebola Treatment Centers (ETC) and from community deaths was established at the entrance of the IPD-ML. On sample receipt an IPD sample request form adapted for EVD suspect cases, containing date of collection, origin, type of samples (serum whole blood, swabs...) was recorded in a register.
Samples processing was subdivided into 3 rooms: (i) room for samples inactivation containing a PSM III glove box (BDK Luft, Genkingen, Germany) or a mobile glove box (Könnecke Präzisionsmechanik, Germany) connected to room (ii) used for viral RNA extraction containing a PSM II (Nuaire, Minnesota, USA) (Figure 1).
Daily first activities included preparation of decontamination solutions (1% Incidin, Ecolab, Germany) and receiving a delivery diesel fuel for the generators which were absoulutely necessary due to the instability of the power supply by the national company, Electricité De Guinée (EDG).
Laboratory staff wore personal protective equipment (PPE), consisting of a protective suit (Tyvek Dupont), a mask, goggles, two pairs of latex gloves with the inner pair taped to the protective suit and a pair shoes and waterproof shoe covers.
On arrival the packaging of samples was disinfected externally by spraying with 1 % incidine and the inner second package transferred into a biosafety cabinet class III (glove box) where specimen were unpacked.
Inactivation of samples was performed inside the glovebox by using the AVL buffer (QIAamp viral RNA mini kit (Qiagen, Hilden, Germany)) as described in the kit guidelines, the sample was additionally incubated for 10 minutes at 60°C and 560µl of Ethanol 70% was added for RNA perciptation.
The tubes were decontaminated externally and moved out of the glove box for into room (ii) for RNA extraction. Extracted RNA was transferred to room (iii). Before leaving room (ii) protective suit, shoe-boots, second pair of gloves were decontaminated with 1% incidine and incinerated.
The third room was divided into 3 sites to prevent carry over contamination: two pipetting sites seperated by aluminium partitions were used for the PCR mix site, and for the positive control site, and the last site was used to add sample RNA to the master mix and to run the real time PCR. The tests were performed by two local technician teams trained during the Ebola outbreak, working 1 day out of 2 under the supervision of 2-3 IPD biologists.
The post-analytical step corresponded to the validation of the results obtained by the technicians. Patient name, age, sex, residence, patient laboratory identifier, collection date, date of symptom onset, clinical status, lab result with Ct value were recorded in an excel database and results dispatched daily to the physician at the ETC, WHO and the national coordination for Ebola response.
Challenges, opportunities and recommendation’s
The availability of a real-time diagnosis allowed rapid diagnosis and supported epidemiologist investigation teams for early detection of cases, isolation, and contact tracing and population awareness.
One issue was the long time spent in collecting and transporting samples inside the isolation wards due to biosafety procedures. This situation affected the rapid care of patients and the implementation of control measures to recover contacts cases after laboratory confirmation.
The lack of adequate material recommended by WHO for the collection and transport of samples lead to the use of inadequate material. Rigorous training for sample receipt avoided any impact on the safety of laboratory staff handling the samples. There was some impact on the quality of the (hemolysis, heparin tubes inhibiting PCR) test results.
However, this epidemic presented also opportunities with international collaboration and exchange between the teams deployed in the field to combat the disease. In addition, the availability of sera bio banks with collected samples will allow the development and evaluation of new diagnostic methods for EBOV.
In terms of recommendation, it is important to strengthen the training of local teams on biosecurity rules and laboratory facilities for Ebola and other hemorrhagic fever diagnosis in affected or endemic areas. It is also crucial to carry out research to develop mobile laboratories which will be used as point of care diagnostic during hemorrhagic fevers outbreaks and thus remove obstacles related to the mobility of field teams in low setting resources. The development of multiplex diagnostic tests is also necessary due to the co-circulation of several pathogens with similar symptoms in affected communities.
Diagnostic activities
The IPD laboratory tested samples from Ebola Treatment Centers (ETC) Médecins sans Frontières at Conakry and the Red Cross at different localities in Guinea. All persons with fever and at least three of the following symptoms: head- aches, lethargy, anorexia/loss of appetite, aching muscles or joints, stomach pain, difficulty swallowing, vomiting, difficulty breathing, diarrhea, hiccups , or fever and contact with known confirmed case were considered as suspect cases. Deceased patients corresponding to the suspect case definition and who had contact with confirmed cases were considered as probable cases. Clinical samples from suspects posed a high risk and were shipped safely to the laboratory (triple packaging) located at the Donka Hospital and handled using class III cabinet. For molecular detection of EBOV, the Realstar® Filovirus screen RT-PCR kit (Altona Diagnostics Hamburg, Germany) was available and recommended by WHO (Altona diagnostic, Filovirus real time RT-PCR). The IPD however used quantitative Taqman RT-PCRs for the detection of hemorrhagic fever viruses including Ebola Zaire, Ebola Sudan, Marburg, Yellow fever, Rift valley fever and Crimean Congo on a portable Smartcycler TD machine developed and validated in the context of the EU project VHF Diagnostics (FP6-INCO-032180) [3-6]. Serological assays were also used for detection of specific IgM and IgG (Winnipeg Canada and CDC USA) for particular cases.
The viral genome can be detected as early as day 1 after disease onset in some patients, and viremia decreased in survivors about 1 week and becomes undetectable about 3 weeks after disease onset [7]. However, EBOV infection cannot be excluded on the basis of a negative EBOV RT-PCR result in the early phase of the disease (1-3 days after the onset of EBOV-associated symptoms in a patient) or beyond a week after disease onset. Therefore, for these particular cases, RT-PCR should be repeated or serological tests should be done to avoid false negative results.
Up to 1st June 2016, 17 623 samples from different regions were tested in the IPD laboratory at Donka Hospital, which corresponded to 55% of all samples tested by the 8 laboratories present in Guinea during the outbreak. In addition to laboratory activities, the IPD team participated in daily meetings organized by the WHO for results reporting.
Research activities
Research activities were also performed by the IPD team during this EVD outbreak. A first study aimed to understand the chains of transmission in order to evaluate and optimize local control strategies for EVD. Confirmed and probable cases of Ebola virus disease reported in Conakry, Boffa, and Télimélé were positioned in chains of transmission. This study showed how one person can spread the disease to his contacts, and identified the community as the major source of infection followed by hospitals and funerals. Implementation of infection control in April 2014 led to reduction of transmission in hospitals and at funerals. In the community, the transmission dropped by 50% for patients that were admitted to hospital, but remained unchanged for those that were not. Simulations analyses showed that a 10% increase in hospital admissions could have reduced the length of chains by 26%. These results supported the objective to increase the capacity of EVD treatment centers both to care for EVD patients and to stop transmission in the community [8].
2021 Copyright OAT. All rights reserv
Another study was done to understand the molecular evolution of EBOV during the outbreak. Phylogenetic analysis revealed circulation of three distinct viral lineages in Guinea, including in the urban setting of Conakry and its surroundings. One lineage was unique to Guinea and closely related to the earliest sampled viruses of the epidemic. A second lineage contained viruses probably reintroduced from neighbouring Sierra Leone on multiple occasions, while a third lineage later spread from Guinea to Mali. Each lineage was defined by multiple mutations, including non-synonymous changes in the virion protein 35 (VP35), glycoprotein (GP) and RNA-dependent RNA polymerase (L) proteins. These data illustrate the ongoing ability of EBOV to develop lineage-specific and potentially phenotypically important variation.
A third study aimed to model case fatality ratios of Ebola patients using viremia [9]. A large dataset of laboratory results for EVD patients in Conakry between March 2014 and February 2015, were analyzed to characterize how viremia can change the probability of death. Viremia was found as an excellent predictor of outcome for individual EVD patients, with the probability of death increasing from 21% in those with low viremia to 81% in the high viremia group. These observations are particularly important for studies that aim to assess the efficacy of treatments in the absence of randomized trials.
Development of rapid test
Implementation of infection control lead to a decrease of transmission in Guinea [8] but reintroductions from Sierra Leone [10] and low community engagement i.e refusal to hospitalize family members, continued unsafe traditional burials, impatience with too long waiting times for test reults, and general sample transportation issues led to prolongation of the outbreak.
Currently, there is no approved vaccine or specific treatment for EVD; therefore, early identification of cases is crucial for the control of EVD epidemics, and a huge challenge in large urban centers.
In this context, the IPD laboratory and collaborators adapted and optimised a rapid test based on isothermal reverse transcription recombinase polymerase amplification (RT-RPA) for the detection of Ebola virus [11,12]. A mobile laboratory consisting of a mobile glove box and a Diagnostics-in-Suitcase (DiaS) powered by a battery and solar panel was used. Before deployment in Conakry a workshop was organized in Dakar, from 19 to 23 January 2015, to evaluate the RPA in laboratory and field conditions without electricity and train Senegalese and Guinean technicians before deployment to Conakry. The efficiency was tested on post mortem swabs in Matoto in Conakry, Guinea as part of the reinforced surveillance strategy in April 2015 to reach the "zero case" goal. A total of 120/928 samples tested positive and the sensitivity and specificity was 100% in reference to real-time RT-PCR and results were provided within 30-60 minutes. This field deployment helped to improve burial management and community engagement.
Roles of IPD laboratory in Senegal
The IPD laboratory played also important roles in Senegal for establishment of preparedness and surveillance. Up to now, the laboratory tested 92 samples from Senegal and confirmed the unique EVD case [13]. Because the WHO recommends that specimens tested by RT-PCR in other laboratories should be sent to a WHO Collaborating Center for confirmatory testing, the first EVD case in Mali was also confirmed in Dakar. The IPD laboratory in Dakar also analyzed suspect cases samples from different African countries, Mali (10), Ghana (1), Bissau Guinea (1), Mauritania (2), Angola (1) and Gambia (4). This highlighted the need of capacity building for EVD diagnostics in Africa. In this context, different workshops were organized in Dakar to strengthen capacities for EVD diagnostic under high bio-safety conditions and prepare a future network response in Africa. From 9 to 12 March 2014, a first workshop was organized in Dakar and twelve African countries (Senegal, Benin, Burkina Faso, Cameroun, Centrafrican Republic, Ivory Coast, Bissau Guinea, Guinea Conakry, Mali, Togo, Tunisia and Morocco) participated in this workshop. During the outbreak, a truck was designed as BSL3+ mobile laboratory by several partners (P4 Lyon, Fondation Merieux, Inserm, Resaolab, Institut Pasteur and Réseau International des Instituts Pasteur). This project called EUWAM-Lab was funded by the EU and the objective was to deploy the mobile laboratory in West Africa for detection of class 4 pathogens including EBOV. For this purpose, African scientists had to be trained on diagnostic tools and bio-safety conditions used in the mobile laboratory. In this context, a second workshop was organized in Dakar from 20 to 24 July 2015 with different countries (France, Senegal, Burkina Faso, Cameroun, Ivory Coast, Guinea Conakry, Mali, Togo, Mauritania and Niger). Currently, the truck is parked in Conakry, Guinea, at Donka Hospital.
On the clinical side, a workshop was also organized in Dakar by the project Inovative Medicines Initiative EbolaMoDRAD (Modern Approaches for developing bedside Rapid Diagnostics). The overall objective of the workshop was to share knowledge on the different aspects of Ebola virus disease: history, clinical diagnosis and interpretation of results, isolation measures, treatment and intensive care, supportive therapy, use of personal protection equipment (PPE), infection prevention and reporting procedures.
Conclusion
This EVD outbreak in West Africa is unprecedented in size and spread since the discovery of the virus in 1976. The mortality rates varied between 28 to 66% depending on the country and the outbreak seriously affected the livelihoods of people and the economies of the affected countries. The outbreak started in December 2013 in Guinea and was only identified and declared as EVD in March 2014 due to a lack of diagnostic tools. This delay in the identification led to the spread of the disease to a significant number of people around the world. In the absence of specific treatments and vaccines, the epidemiological surveillance and laboratory played crucial roles for the confirmation of suspect cases and release of non-cases during the outbreak. Re-introductions of the virus from survivors were observed in Liberia, and this highlighted the important roles of surveillance and laboratory for rapid detection of any reintroduction or re-emergence of EVD.
The IPD laboratory is a WHO-approved collaborating Centre and was the first deployed laboratory to Conakry after the declaration of the disease. The IPD laboratory played important roles in (1) diagnostics in Guinea and tested 55% of all samples in the country, (2) better understanding of transmission chains in Conakry and the molecular evolution of the virus during the outbreak and (3) improvement of burial management and community engagement by developing a rapid test.
It is uncertain when and where the next EVD outbreak may occur. Therefore, it is crucial for all countries at risk, to be prepared for detection of the disease as early as possible, and to have appropriate responses. In this context, it is crucial to strengthen capacities and the IPD laboratory organized different workshops for capacity building and network response for future.
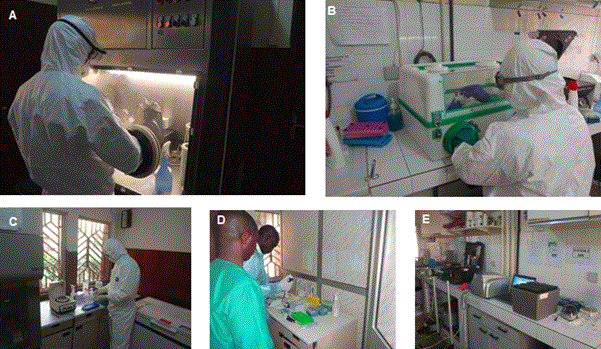
Figure 1. Laboratory set up. (A-B) room for sample inactivation, glovebox in B used in case of power failure and in field deployment, (C) room for RNA extraction, (D) Room for master mix preparation (E) room to add sample RNA to the master mix and to run the real time PCR.
Acknowledgements
Welcome Trust programme: Research for Health in Humanitarian Crises (R2HC) 13376: Point-of-care diagnostic testing for Ebola virus disease in Ebola treatment centres.
References
- Ebola virus disease in Guinea. Cited from: http://www.who.int/csr/don/2014_03_23_ebola/en/.
- Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, et al. (2014) Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med 371: 1418-1425. [Crossref]
- Weidmann M, Muhlberger E, Hufert FT (2004) Rapid detection protocol for filoviruses. J Clin Virol 30: 94-99.
- Weidmann M, Hufert FT, Sall AA (2007) Viral load among patients infected with Marburgvirus in Angola. J Clin Virol 39: 65-66. [Crossref]
- Weidmann M, Sanchez-Seco MP, Sall AA, Ly PO, Thiongane Y, et al. (2008) Rapid detection of important human pathogenic Phleboviruses. J Clin Virol 41: 138-142.
- Weidmann M, Faye O, Faye O, Kranaster R, Marx A, et al. (2010) Improved LNA probe-based assay for the detection of African and South American yellow fever virus strains. J Clin Virol 48: 187-192. [Crossref]
- Vernet MA, Reynard S, Fizet A, Schaeffer J, Pannetier D, et al. (2017) Clinical, virological, and biological parameters associated with outcomes of Ebola virus infection in Macenta, Guinea. JCI Insight 2: e88864. [Crossref]
- Faye O, Boelle PY, Heleze E, Faye O, Loucoubar C, et al. (2015) Chains of transmission and control of Ebola virus disease in Conakry, Guinea, in 2014: an observational study. Lancet Infect Dis 15: 320-326. [Crossref]
- Faye O, Andronico A, Faye O, Salje H, et al. (2015) Use of Viremia to Evaluate the Baseline Case Fatality Ratio of Ebola Virus Disease and Inform Treatment Studies: A Retrospective Cohort Study. PLoS Med 12: e1001908. [Crossref]
- Simon-Loriere E, Faye O, Faye O, Koivogui L, Magassouba N, et al. (2015) Distinct lineages of Ebola virus in Guinea during the 2014 West African epidemic. Nature 524: 102-104. [Crossref]
- Euler M, Wang Y, Heidenreich D, Patel P, Strohmeier O, et al. (2013) Development of a Panel of Recombinase Polymerase Amplification Assays for Detection of Biothreat Agents. J Clin Microbiol 51: 1110-1117. [Crossref]
- Faye O, Faye O, Soropogui B, Patel P, El Wahed AA, et al. (2015) Development and deployment of a rapid recombinase polymerase amplification Ebola virus detection assay in Guinea in 2015. Euro Surveill 20. [Crossref]
- Ka D, Fall G, Diallo VC, Faye O, Fortes LD, et al. (2017) Ebola Virus Imported from Guinea to Senegal, 2014. Emerg Infect Dis 23: 1026-1028. [Crossref]

