Abstract
Diabetes has turned out to be pandemic towards the end of the century; unfortunately there are many people who are still unaware of the fact that they are having diabetes even if they have already developed various complications related to it. Patients with long duration of Type 2 Diabetes Mellitus have a significantly increased risk of developing related complications. This ignorance is due to similar manifestations for two related health problems like that of heart problem which may or may not be related to diabetes. Finding genetic variants in genes is a pretty good method for assessment of the risk for type 2 diabetes mellitus and the complications related to it. These genetic predisposing factors can foretell the susceptibility of a person to the disease or become the reason behind diabetes in the person. The present case control study intends to associate the reported single nucleotide polymorphisms in seven genes viz. Calpain10 (CAPN10), Adiponectin (ADIPOQ), Apolipoprotein C-III (APOC III)], Angiotensinogen (AGT), Apolipoprotein E (APOE) and Toll-Like Receptor 4(TLR4) genes as risk factors for Type 2 Diabetes and its related Complications in the discrete diabetic population of Andhra Pradesh and Telangana states in India. The results showed the association some of the single nucleotide polymorphisms in the candidate genes which increased the risk of development of type 2 diabetes mellitus and progression in the form of complications.
The results showed the presence of new Single Nucleotide Polymorphism in the Type 2 Diabetes cases. Further extension of analyses work on a larger sample size is to be undertaken in our study population.
Key words
Type 2 diabetes mellitus (T2DM), complications, predisposing factors, genes, new Single Nucleotide Polymorphisms (SNPs), genetic variants
Introduction
Diabetes mellitus is a heterogeneous group of metabolic disorder characterized by dysregulated glucose metabolism. Its development and advancement in form of complications are due to several risk factors. Uncontrolled Diabetes or elevated blood glucose levels for prolonged time lead to long term and short term complications like [1]microvascular (damage to small blood vessels) and macrovascular (due to damage to the arteries). The high blood glucose levels for prolonged time affect both the quality and length of life [2,3]. This condition is attributed to impairment of glucose utilization. The insulin resistance [4,5] or deficiency [6] is responsible for this condition. The prevalence of T2DM is increasing on daily basis in most of the world wide countries [7] with a significant effect on the adult population [8]. It is expected to rise to 529 million by 2035 (Diabetes Atlas-update-2014). The increase is more in especially developing countries [3] like India. India is designated as “Diabetic capital of the World” for it has the largest increase of 80% diabetic subjects than any other country. (The International Diabetes Federation) The Characteristic “Asian Indian Phenotype” makes them more susceptible to the disease [9]. Among them the people living in southern states are more prone due to their life style [10]. The people who have well-controlled blood sugar levels have less common and less severe in complications of diabetes mellitus [11]. Several risk factors can foretell the susceptibility of a person to the type 2 diabetes mellitus and its related complications. They are grouped into Genetic, Environmental and Biochemical risk factors [12]. This factors either individually or on interaction with one another to cause the disease. The structural variations in the genes are responsible for variation in phenotype [13,14] is known to be the genetic risk factors.
Calpain 10 (CAPN10)
Calpain10 gene [15,16] encoded by cysteine protease is the first but not the last candidate gene with susceptibility for T2DM. It was identified through a positional cloning and genome wide screening [17]. Its role regulation of the glucose is known. Studies on CAPN 10 for T2DM among different ethnic groups in wide range of population showed uneven results [18-23] due to genetic heterogeneity between populations. Some confirming the original finding that haplotype combination comprising three intronic CAPN10 single-nucleotide polymorphisms (UCSNP-43, -19, and -63) was associated with increased risk of T2DM but some do not.
Adiponectin (ADIPOQ)
ADIPOQ gene is located on Chromosome 3q27 is responsible for the major adipocyte secretory protein. It has a potent role in the metabolism of fat and the regulation of glucose. It includes insulin sensitivity in muscle and liver, regulating energy homeostasis and glucose tolerance. The low circulating levels of adiponectin are associated with obesity and diabetes [24,25].
Dickkopf WNT signaling pathway inhibitor 1(DKK1)
This gene encodes a cysteine rich glycoproteins and is a member of the dickkopf family. Elevated levelsof DKK1 in bone marrow plasma and peripheral blood are associated with the presence of osteolytic bone lesions .It is involved in glucose metabolism [26].
Apolipoprotein C-III (APOC III)
Apolipoproetin is the product of this gene. It induces the development of hypertriglyceridemia by delaying the catabolism of triglyceride-rich particles [27].
Angiotensinogen (AGT)
This gene belongs to a family of SERPIN. This produces protein angiotensin protein that is part of the renin-angiotensin system that regulates blood pressure [28].
Apolipoprotein E (APOE)
The APOE gene provides instructions for making a protein called apolipoprotein E. It mediates the binding, internalization, and catabolism of lipoprotein particles [29].
Toll-Like Receptor 4(TLR4)
The protein encoded by this gene is a member of the Toll-like receptor (TLR) family which plays a fundamental role in the diseases associated with protein coding TLR4 include endotoxin hypo responsiveness and macular degeneration [30].
Materials and methods
Enrolment of cases and controls
The discrete population of Andhra Pradesh and Telangana states in India was selected for the case control study. For this a total of 200 individuals visiting diagnostic centres were enrolled. They were primarily screened for diabetes using glucose oxidase peroxidase method [31] in accordance to WHO criteria. Out of which a total of 180 individuals (96 females and 84 males) of 15-85 years of age were selected after the exclusion of those with other serious disorders.Those with Fasting Blood Sugar (FBS) value of more than 126 mg/dl or ≥ 7.0 mmol/L and Post Prandial Blood Sugar (PPBS) value of more than 200 mg/dl or ≥ 11.1mmol/L (WHO diagnostic criteria for diabetes) are cases and those with no family history of diabetes with FBS and PPBS value less than above values served as controls.
Collection of the Blood Sample
Before the collection of sample the subjects of the study were subjected to simple questionnaire for the details of family history, habits, and duration of diabetes before the procurement of blood sample. Thereafter in accordance to protocol 3ml of the left over blood samples was collected with 5ml syringe in the EDTA [32] vacutainers for the SNP study. A total of 51 samples (41 diabetic cases and 10 controls) were processed for all the seven genes.
Selection of polymorphism
We limited our selection of genetic variants to the previously reported Single nucleotide polymorphisms in recent epidemiological studies for all the seven candidate genes for T2DM and its related complications at the time of initiation of our studies. We selected a set of SNPs with reference sequence rs2975760 [33] and rs3792267 [34] in the CAPN10, and rs3774261 [35] in ADIPOQ for T2DM, rs1501299 [35] and rs2241767 [35] in ADIPOQ. The other SNPs of candidate genes with proven association with T2DM complications are rs1569198 [36] in DKK1for Musculoskeletal Disorders, rs121918381 [37] in APOC III for Dyslipidemia; rs699 [38] in AGT for Hypertension; rs429358 [39] in APOE for Alzheimer’s disease [40] and rs1927911 [41] in TLR 4 for diabetic Foot Ulcer were selected.
Genotyping
The Leucocyte component of the blood samples were processed for extraction and purification of Genomic DNA using modified Sambrook et al protocol [42,43]. The isolated and purified DNA was amplified in single nucleotide Polymorphism specified region by the Polymerase chain reaction [44,45] using specially designed primers and synthesized by synthesized by Bioserve, Hyderabad, India. The primers were designed by using Primer blast tool [44]. The PCR conditions were optimized to get the maximum yields of target DNA sequences. PCR was performed with reaction mixture of 20μl volume. The reaction mixture consists of the primers, (forward and reverse), Taq polymerase enzyme, reaction buffer, Master mix (dNTP, MgCl2), DNA and water. The ethidium bromide stained amplified product of PCR was qualitatively checked on 1.2% agarose gel electrophoresis [36,37]. The gel was visualized and photographed in a transilluminator (Figure 1).
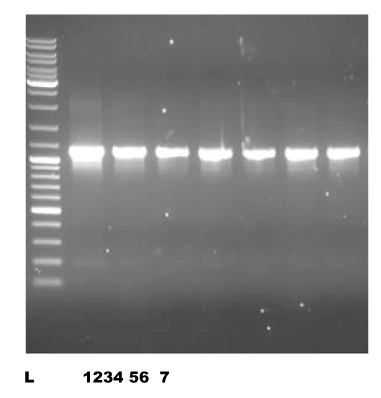
Figure 1. Photographic image of PCR product on Agarose Gel
(L= MW Ladder, 1=CAPN10,2=ADIPOQ,3=DKK1, 4=AGT,5=APOC III ,6=APOE,7= TLR4 DNA Samples)
The amplicons were then sequenced using Sanger sequencing method [38]. Mutations in all the genes were screened by different computational tools. They were checked for the validation of mutations in our population.
Results
The results showed difference in mean and standard deviation of Fasting and post-prandial blood glucose levels in T2DM cases and controls. The values for blood glucose are shown in Table1. The graphical representation of the Blood sugar values is shown in Graph 1and Graph 2.
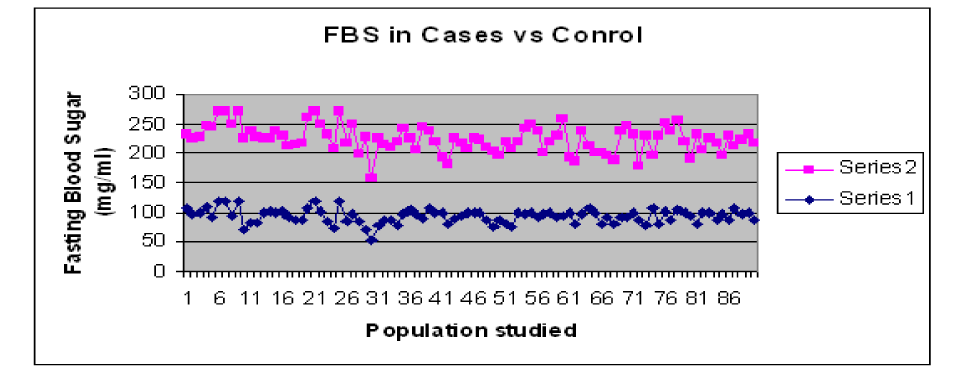
Graph 1. Comparison of FBS levels in T2DM Cases and Control
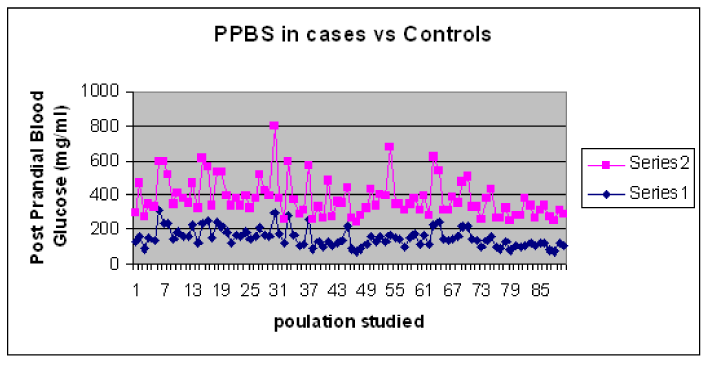
Graph 2. Comparison of PPBS levels in T2DM Cases and Control
Parameter |
T2DM CASES n=90 |
CONTROLS n=90 |
Normal Range mg/dl |
P value |
FBS |
152 ± 92.8 |
92 ± 11.7 |
70-110 |
0.8 |
PPBS |
229.3 ± 67.6 |
131.2 ± 18.8 |
170-200 |
0.7 |
Table 1. Fasting and Postprandial Blood sugar Levels
The sequencing of amplified portion of the two genes revealed the presence of previously reported SNPs in the diabetic population are shown below in the Table 2. The SNPs rs2975760, rs3792267 in CAPN10 and rs3774261 ADIPOQ genes are reported for diabetes are present in 48.7% and 41.4% respectively. The previously SNPs reported for complications are present in 43.9% and 46.3% in (rs1501299, rs2241767)ADIPOQ, 63.41% in (rs1569198 )DKK1, 43.9% in (rs121918381)APOC III, 48.7% in AGT, 60.9% in (rs429358)APOE, 31.7% in (rs1927911)TLR4.
GENE |
Reported SNP |
Reported SNP type |
Amplicon sequence |
% of subjects with reported SNP |
New SNP details
(Position) |
CAPN10
|
rs2975760
rs3792267 |
C/T
A/G |
240591700– 240592223
|
48.7%
48.7%
31.7% |
240592222 |
ADIPOQ |
rs3774261
rs1501299
rs2241767 |
A/G
A/C(REV)
A/G
|
186853309- 186853804 |
41.4%
43.9%
46.3% |
|
DKK1 |
rs1569198 |
A/G |
52316013 - 52316520 |
63.41%
48.7% |
52316022 |
APOC III |
rs121918381 |
A/G |
116832368-116832880 |
43.9% |
|
AGT |
rs699
|
C/T(REV)
|
230710528- 230710030
(REV) |
48.7% |
|
APOE |
rs429358 |
C/T 44908684 |
44908662 -44909173 |
60.9% |
|
TLR4 |
rs1927911 |
C/T 11707776
G/A |
11707736 - 11708239 |
31.7%
21.7% |
11708205 |
Table 2. Distribution of reported and Novel SNP in Diabetic cases
The BOX SHADE (Table 3) of the amplified gene shows the presence of SNPs in new position in CAPN10, DKK1 and TLR4 genes along with the previously reported SNPs in south Indian population. Only the SNP reported in DKK1 in the world wide population could not be found in any of the test samples.
CAPN10
|
DKK1 |
TLR4 |

240592222 |
|
52316022

11708205 |
Table 3. BOX SHADE showing Position of Novel SNPs
Conclusion
The association of the reported SNPs for diabetes and related complications are successfully replicated in our discrete study population. A normal individual carrying these gene variants may face a greater risk of developing diabetes if having uncontrolled blood sugar levels that exceeds the values of WHO recommendation criterion for diabetes diagnosis.
We conclude that genetic variation in the genes influences blood glucose levels in making non-diabetic subjects susceptible to diabetes and its progression to complications This shows the combined effect of more than one factor for outcome of diabetes.
The SNP found in new position could be new biomarkers for type 2 diabetes mellitus. However, further analyses on a larger sample size are required to establish a conclusive association of the novel SNPs with diabetes and its related complications to diabetic population of Andhra Pradesh and Telangana states in India.
Acknowledgements
We humbly acknowledge all the participants of the study.
References
- Kitabchi AE, Umpierrez GE, Miles JM, Fisher JN (2009) Hyperglycemic crises in adult patients with diabetes. Diabetes Care 32: 1335-1343. [Crossref]
- Diabetes mellitus – Wikipedia Acessed on 5th Feb 2016
- Twito O, Frankel M, Nabriski D (2015) Impact of glucose level on morbidity and mortality in elderly with diabetes and pre-diabetes. World J Diabetes 6: 345-351.
- O'Doherty R, Stein D, Foley J (1997) Insulin resistance. Diabetologia 40 Suppl 3: B10-15. [Crossref]\
- Lillioja S, Mott DM, Spraul M, Ferraro R, Foley JE, et al. (1993) Insulin resistance and insulin secretory dysfunction as precursors of non-insulindependent diabetes mellitus. Prospective studies of Pima Indians. N Engl J Med 329: 1988-1992. [Crossref]
- Temple RC, Carrington CA, Luzio SD, Owens DR, Schneider AE, et al. (1989) Insulin deficiency in non-insulin-dependent diabetes. Lancet 1: 293-295. [Crossref]
- King H, Aubert RE, Herman WH (1998) Global burden of diabetes, 1995-2025: prevalence, numerical estimates, and projections. Diabetes Care 21: 1414-1431. [Crossref]
- Wild S, Roglic G, Green A, Sicree R, King H (2004) Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 27: 1047-1053. [Crossref]
- Mohan V, Sandeep S, Deepa R, Shah B, Varghese C (2007) Epidemiology of type 2 diabetes: Indian scenario. Indian J Med Res 125: 217-230. [Crossref]
- Mohan V, Pradeepa R (2009) Epidemiology of diabetes in different regions of india. Health Administrator XXII: 1-18.
- "Diabetes Complications". Diabetes.co.uk. Retrieved 2 February 2016.
- Cheekurthy AJ, Rambabu C, Kumar A, Surendrababu K (2015) Predisposition factors of T II Diabetes Mellitus and related complications, Biotechnology and Bioforensics. Springer Briefs in Applied Sciences and Technology 5: 43-50.
- Shastry BS (2009) SNPs: impact on gene function and phenotype. Methods Mol Biol 578: 3-22. [Crossref]
- Barøy T, Misceo D, Frengen E (2008) [Structural variation in the human genome contributes to variation of traits]. Tidsskr Nor Laegeforen 128: 1951-1955. [Crossref]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, et al. (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26: 163-175.
- Hoffstedt J, Rydén M, Löfgren P, Orho-Melander M, Groop L, et al. (2002) Polymorphism in the Calpain 10 gene influences glucose metabolism in human fat cells. Diabetologia 45: 276-282. [Crossref]
- Ridderstrale M, Nilsson E (2008) Type 2 diabetes candidate gene CAPN10: first, but not last. Curr Hypertens Rep 10: 19-24. [Crossref]
- Fullerton SM, Bartoszewicz A, Ybazeta G, Horikawa Y, Bell GI, et al. (2002) Geographic and haplotype structure of candidate type 2 diabetes susceptibility variants at the calpain-10 locus. Am J Hum Genet 70: 1096-1106. [Crossref]
- Lynn S, Evans JC, White C, Frayling TM, Hattersley AT, et al. (2002) Variation in the calpain-10 gene affects blood glucose levels in the British population. Diabetes 51: 247-250. [Crossref]
- Rasmussen SK, Urhammer SA, Berglund L, Jensen JN, Hansen L, et al. (2002). Variants within the calpain-10 gene on chromosome 2q37 (NIDDM1) and relationships to type 2 diabetes, insulin resistance, and impaired acute insulin secretion among Scandinavian Caucasians. Diabetes 51: 3561-3567. [Crossref]
- Elbein SC, Chu W, Ren Q, Hemphill C, Schay J, et al. (2002) Role of calpain-10 gene variants in familial type 2 diabetes in Caucasians. J Clin Endocrinol Metab 87: 650-654. [Crossref]
- Adak S, Sengupta S, Chowdhury S, Bhattacharyya M (2010) Co-existence of risk and protective haplotypes of Calpain 10 gene to type 2 diabetes in the eastern Indian population. Diab Vasc Dis Res 7: 63-68. [Crossref]
- Sharma R, Matharoo K, Kapoor R, Chopra H, Bhanwer AJ (2013) Ethnic differences in CAPN10 SNP-19 in type 2 diabetes: a North-West Indian case control study and evidence from meta-analysis. Genet Res (Camb) 95: 146-155. [Crossref]
- Matharoo K, Arora P, Bhanwer AJ (2013) Association of adiponectin (AdipoQ) and sulphonylurea receptor (ABCC8) gene polymorphisms with Type 2 Diabetes in North Indian population of Punjab. Gene 527: 228-234. [Crossref]
- Chu H, Wang M, Zhong D, Shi D, Ma L, et al. (2013) AdipoQ polymorphisms are associated with type 2 diabetes mellitus: a meta-analysis study. Diabetes Metab Res Rev 29: 532-545. [Crossref]
- Jin T (2008) The WNT signalling pathway and diabetes mellitus. Diabetologia 51: 1771-1780. [Crossref]
- Ginsberg HN, Brown WV (2011) Apolipoprotein CIII: 42 Years Old and Even More Interesting. Arterioscler Thromb Vasc Biol 31: 471-473. [Crossref]
- Sonna LA, Glueck SB, Jeunemaître X (2002) Exercise, genetics, and blood pressure: focus on "physical exercise and blood pressure with reference to the angiotensinogen M235T polymorphism" and on "angiotensinogen M235T polymorphism associates with exercise hemodynamics in postmenopausal women". Physiol Genomics 10: 45-47. [Crossref]
- Liu CC, Kanekiyo T, Xu H, Bu G (2013) Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 9: 106-118. [Crossref]
- Visintin A, Mazzoni A, Spitzer JH, Wyllie DH, Dower SK, et al. (2001) Regulation of Toll-like receptors in human monocytes and dendritic cells. J Immunol 166: 249-255. [Crossref]
- Huggett AS, Nixon DA (1957) Use of glucose oxidase, peroxidase, and O-dianisidine in determination of blood and urinary glucose. Lancet 273: 368-370. [Crossref]
- Banfi G, Salvagno GL, Lippi G (2007) The role of ethylenediamine tetraacetic acid (EDTA) as in vitro anticoagulant for diagnostic purposes. Clin Chem Lab Med 45: 565-576. [Crossref]
- Khan IA, Movva S, Shaik NA, Chava S, Jahan P, et al. (2014) Investigation of Calpain 10 (rs2975760) gene polymorphism in Asian Indians with Gestational Diabetes Mellitus. Meta Gene 2: 299-306. [Crossref]
- Kommoju UJ, Maruda J, Kadarkarai Samy S, Irgam K, Kotla JP, et al. (2014) Association of IRS1, CAPN10, and PPARG gene polymorphisms with type 2 diabetes mellitus in the high-risk population of Hyderabad, India. J Diabetes 6: 564-573. [Crossref]
- Ramya K, Ayyappa KA, Ghosh S, Mohan V, Radha V (2013) Genetic association of ADIPOQ gene variants with type 2 diabetes, obesity and serum adiponectin levels in south Indian population. Gene 532: 253-262. [Crossref]
- Piters E, Balemans W, Nielsen TL, Andersen M, Boudin E, et al. (2010) Common genetic variation in the DKK1 gene is associated with hip axis length but not with bone mineral density and bone turnover markers in young adult men: results from the Odense Androgen Study. Calcif Tissue Int 86: 271-281. [Crossref]
- The TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute (2014) Loss-of-Function Mutations in APOC3, Triglycerides, and Coronary Disease. NEJM 371: 22-31.
- Karthikeyan M, Shridevi V, Rose R, Anandan B, Singh KD, et al. (2013) Angiotensin Gene Polymorphisms (T174M and M235T) are SignificantlyAssociated with the Hypertensive Patients of Tamil Nadu, South India. Int J Hum Genet 13: 201-207.
- Mohana VU, Swapna N, Surender RS, Vishnupriya S, Padma T (2012) Gender-related association of AGT gene variants (M235T and T174M) with essential hypertension--a case-control study. Clin Exp Hypertens 34: 38-44. [Crossref]
- Prada D, Colicino E, Power MC, Cox DG, Weisskopf MG, et al. (2014) Influence of multiple APOE genetic variants on cognitive function in a cohort of older men results from the Normative Aging Study. BMC Psychiatry 14: 223. [Crossref]
- Singh K, Singh VK, Agrawal NK, Gupta SK, Singh K (2013) Association of Toll-Like Receptor 4 Polymorphisms with Diabetic Foot Ulcers and Application of Artificial Neural Network in DFU Risk Assessment in Type 2 Diabetes Patients. BioMed Research International.
- Sambrook J, Green MR (1989) Molecular Cloning: A Laboratory Manual, ColdSpring Harbor Laboratory Press, New York.
- Subbarayan PR, Sarkar M, Ardalan B (2002) Isolation of genomic DNA from human whole blood. Biotechniques 33: 1231, 1234. [Crossref]
- Mullis KB, Faloona FA (1987) Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol 155: 335-350. [Crossref]
- Mullis KB (1990) The unusual origin of the polymerase chain reaction. Sci Am 262: 56-61, 64-5. [Crossref]
- Stellwagen NC (2009) Electrophoresis of DNA in agarose gels, polyacrylamide gels and in free solution. Electrophoresis 30 Suppl 1: S188-195. [Crossref]
- Johnson PH, Grossman LI (1977) Electrophoresis of DNA in agarose gels.Optimizing separations of conformational isomers of double- and singlestranded DNAs. Biochemistry 16: 4217-4225. [Crossref]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74: 5463-5467. [Crossref]