Abstract
Aims: To determine the differential profile of microRNA (miRNA) in the aqueous humor (AH) of patients with primary congenital glaucoma (PCG) when compared to AH from children with congenital cataract.
Methods: AH samples were obtained during surgery from patients with PCG (n= 6) and congenital cataract (n=6) that served as controls. Following miRNA extraction from ~100 µL of AH, the presence of small RNAs (<200 nucleotides) was confirmed using a bioanalyzer (Agilent). Labelled miRNA was then hybridized to miRNA microarray 4.0 (Affymetrix) according to the standard protocol. The miRNA microarray data was analyzed using Partek Genomic Suite 6.13 and one-way ANOVA statistical testing to identify significantly differential expressed miRNAs. Pathway analysis was performed using DIANA tools (mirPath) to identify pathways associated with differential expressed (DE) miRNAs.
Results: A total of 2578 miRNAs was quantitated in AH samples. Analysis revealed 11 DE miRNA in the AH from PCG patients. mirPath identified the Wnt signaling pathway as the major pathway associated with 5 DE miRNAs [hsa-miR-4659a-5p, hsa-miR-548aa, hsa-miR-548t-3p, hsa-miR-606 and hsa-miR-548aj-3p that correlated with 5 genes: ROCK1 (ENSG00000067900), SMAD2 (ENSG00000175387), NFAT5 (ENSG00000102908), PRKCB (ENSG00000166501) and TBL1XR1 (ENSG00000177565), respectively.
Conclusion: The AH from patients with PCG shows a differential miRNA profile when compared to a congenital cataract control group. Most of the specific miRNA’s that were down-regulated are associated with the Wnt signaling pathway and may be involved in trabecular meshwork dysfunction, similar to that reported in adult glaucoma.
Key words
congenital glaucoma, genetics, miRNA, Wnt signaling
Introduction
Primary congenital glaucoma (PCG; OMIM 231300) is an autosomal recessive form of glaucoma that manifests itself in infancy and is characterized by developmental defect(s) of the trabecular meshwork (TM) and anterior chamber angle affecting the aqueous humor (AH) outflow pathway [1]. CYP1B1 (OMIM *601771; cytochrome P450, subfamily I, polypeptide I) mutations are the predominant cause of PCG in humans [2]. Although a recent animal study supports an oxidative stress-induced model of PCG, the precise mechanism(s) by which CYP1B1 regulates development of the anterior chamber remains elusive [3]. LTBP2 (OMIM *602091; latent transforming growth factor beta binding protein 2) mutation as a cause of PCG is extremely rare; being reported only in a few families from Pakistan, Iran, and European gypsies [4]. It is thus evident that other genes/proteins may be altered in PCG and are still to be identified.
Alteration in the composition of AH proteins has been associated with several eye diseases including PCG [5]. Recent studies have provided evidence for the involvement of molecular mediators such as microRNAs (miRNAs) in the pathophysiology of glaucoma [6]. Furthermore, the identification of miRNAs in ocular cells[7] AH of patients with cataract [8], exosomes isolated from AH [9] and vitreous fluid [10] suggests that miRNAs may have roles to play in the function of the eye and eye diseases.
Many studies have reported miRNA dysregulation in several ocular diseases. These include differential miRNA expression in central epithelium of transparent and age-related cataractous human lenses [11]; in primary human Tenon's fibroblasts treated with transforming growth factor beta 1 (TGFβ1) [12]; in AH of patients with glaucoma[13]; and formalin fixed paraffin embedded tissue samples of patients with intraocular medulloepithelioma [14].
The aim of this study was to evaluate miRNA profiles in AH of patients with PCG and compare them with miRNA profiles in the AH of patients with congenital cataract (serving as control group).
Materials and methods
AH of participants with PCG (n=6) serving as the patient group and congenital cataract (n=6) used as a comparative control group were examined for differential miRNA expression. All PCG patients had confirmed clinical diagnosis for PCG and were all positive for CYP1B1 mutation (G61E) common among the Saudi PCG patients. Patients with previous surgery in affected eye, ocular inflammation and syndrome congenital glaucoma were excluded from the study. Approximately 100-200 µL of AH was withdrawn through a paracentesis track prior to initiation of glaucoma cataract surgery. The samples were immediately centrifuged for 10 mins at 10,000xg to remove cellular debris. miRNA was extracted with Qiagen miRNeasy mini-kit (Qiagen Inc. , Valencia, CA, USA) from approximately 100 µL of AH collected from each patient using the supplementary Protocol:RY43 Purification of total RNA including small RNA from serum or plasma. The presence of small RNAs (<200 nucleotides) was confirmed by Agilent 2100 bioanalyzer (Agilent Technologies, Inc. Santa Clara, CA, USA) using an Agilent Bioanalyzer Pico Chip. miRNAs from individual AH samples were amplified using Ncode miRNA amplification system v.2 (Life Technologies, Grand Island, NY, USA). A total of 130 ng of amplified miRNA was used for labelling with FlashTag HSR labelling kit (Affymetrix, Santa Clara, CA, USA) with the modification of omitting A tailing step as the amplified miRNA is already A tailed during amplification step. The labelled miRNA was then hybridized to Affymetrix miRNA array 4.0 according to the standard protocol. Affymetrix miRNA 4.0 is the newest miRNA array from Affymetrix and it provides 100% miRBase v20 coverage including 2578 human mature miRNA probe sets, 1996 human snoRNA and scaRNA probe sets, and 2025 human pre-miRNA probe sets. Robust Multiarray Averaging (RMA) was performed and the miRNA microarray data was analyzed using Partek Genomic Suite 6.13 and one-way ANOVA statistical testing was performed to identify significantly differential expressed miRNA between different groups (P ≤ 0.05). The differential miRNAs identified were then analyzed with DIANA tools (mirPath) available at (http://diana.imis.athena-innovation.gr/DianaTools/index.php) [15]. mirPath is designed to highlight pathways associated with miRNAs identified. Next, we utilized an option within mirPath called "pathway intersection". This option provides the intersection of targeted pathways by the selected miRNAs. The resulting subset contains only pathways with statistically significant results for all the selected miRNAs.
This study was approved by the King Khaled Eye Specialist Hospital Institutional Review Board and followed the principles established in the Declaration of Helsinki. Written informed consent was obtained from all study participants and all samples were anonymized to preserve patient confidentiality.
Results
The expression of 2578 miRNAs was quantitated in AH from patients with PCG (patient group) and congenital cataract (control group). The demographic information and clinical profile of both study groups is summarized in Table 1.
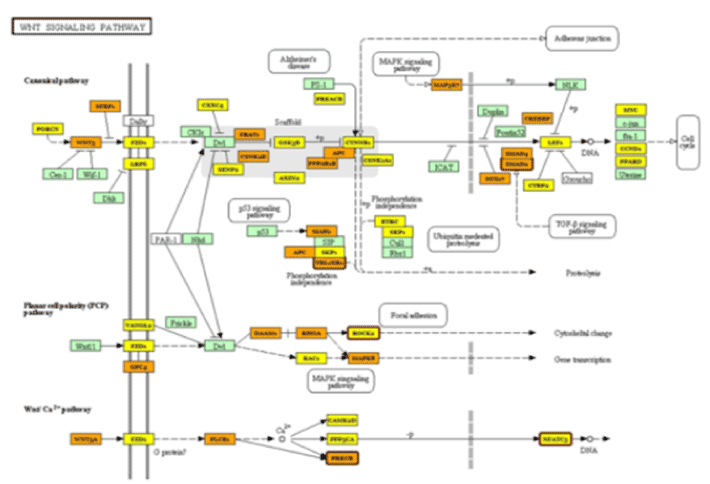
Figure 1. Wnt signaling pathway
Table 1. Demographic and Clinical profile of subjects based on diagnosis
Variables |
Primary congenital glaucoma
(n = 6) |
Congenital cataract
(n = 6) |
P-value† |
Age in years |
|
|
|
Median |
3.5 |
5 |
- |
25% quartile |
2.0 |
3.75 |
- |
Range |
2 - 11 |
3 - 10 |
- |
Intraocular pressure in mmHg,
Mean (SD) |
28.3 (11.6) |
17.5 (2.1) |
- |
Gender, No. (%) |
|
|
|
Male |
4 (66.7) |
6 (100) |
- |
Female |
2 (33.3) |
0 (0) |
0.45 |
No. of glaucoma medications (%) |
|
|
|
No medication |
0 (0) |
6 (100) |
- |
1 - 2 |
4 (66.7) |
0 (0) |
0.0047 |
>2 |
2 (33.7) |
0 (0) |
0.035 |
Previous surgery |
|
|
|
No |
1 (12.7) |
0 (0) |
- |
Yes |
5 (83.3) |
6 (100) |
0.99 |
†Fishers’ exact probability test
Comparison of miRNA profile in PCG group vs. controls revealed 11 differentially expressed miRNAs (Table 2) [P ≤ 0.05, using one-way ANOVA]. Five (45.5%) of those miRNAs were up-regulated with fold change ranging from +1.12 to 2.30, whereas 6 (54.5%) miRNAs were down-regulated with fold change ranging from (- 1.35 to - 2.26).
Table 2. Fold change of differentially expressed aqueous humor miRNAs in patients with primary congenital glaucoma as compared to controls
DE miRNAs |
Fold Change
in PCG |
P-value |
hsa-miR-4659a-5p |
-1.35 |
0.010125 |
hsa-miR-4445-3p |
+2.20 |
0.020018 |
hsa-miR-3201 |
+1.56 |
0.0403058 |
hsa-miR-548aa |
-2.26 |
0.0214654 |
hsa-miR-548t-3p |
-2.26 |
0.0214654 |
hsa-miR-3128 |
-1.88 |
0.0314196 |
hsa-miR-4423-3p |
-1.64 |
0.020099 |
hsa-miR-606 |
-1.41 |
0.0342179 |
hsa-miR-638 |
+2.30 |
0.0219638 |
hsa-miR-548aj-3p |
+1.12 |
0.00974459 |
hsa-miR-6777-5p |
+1.28 |
0.0148227 |
miRNA in bold are the ones involved in Wnt Signaling Pathway.
Negative (-) sign indicates down-regulation and (+) sign indicates up-regulation. Five of the eleven miRNAs were up-regulated and six were down-regulated in PCG patients.
Analysis of 11 differentially expressed miRNA associated with patient group (PCG) using the DIANA tools (mirPath) identified 63 pathways related to signaling, metabolism and cancer (Table 3). Further analysis using the "pathway intersection" tool within mirPath identified one pathway (Wnt Signaling pathway) involving 5 miRNAs [hsa-miR-4659a-5p, hsa-miR-548aa, hsa-miR-548t-3p, hsa-miR-606 and hsa-miR-548aj-3p that correlated with 5 genes: ROCK1 (ENSG00000067900), SMAD2 (ENSG00000175387), NFAT5 (ENSG00000102908), PRKCB (ENSG00000166501) and TBL1XR1 (ENSG00000177565)], respectively (Figure 1). Four of these miRNAs were down-regulated and one was up-regulated. Due to lack of sufficient AH, the differential expression of these miRNAs could not be validated by a secondary quantitative real-time PCR analysis. Regardless, Figure 1 clearly demonstrates that these differentially expressed miRNAs significantly impact Wnt signaling pathways. Table 4 indicates their possible role in Wnt modulation, however, further work is needed to understand the manifestation and implication of these links in the context of PCG.
Table 3. Pathways identified
Pathway identified |
#miRNAs |
Ras signaling pathway |
9 |
Glutamatergic synapse |
9 |
Dopaminergic synapse |
9 |
Amphetamine addiction |
9 |
cAMP signaling pathway |
9 |
TGF-beta signaling pathway |
8 |
Signaling pathways regulating pluripotency of stem cells |
8 |
Ubiquitin mediated proteolysis |
8 |
Focal adhesion |
8 |
Long-term potentiation |
8 |
Viral carcinogenesis |
8 |
Protein processing in endoplasmic reticulum |
8 |
Osteoclast differentiation |
8 |
PI3K-Akt signaling pathway |
8 |
Cholinergic synapse |
8 |
Hippo signaling pathway |
7 |
Proteoglycans in cancer |
7 |
Adherens junction |
7 |
Transcriptional misregulation in cancer |
7 |
Wnt signaling pathway |
7 |
Prolactin signaling pathway |
7 |
Sphingolipid signaling pathway |
7 |
Adrenergic signaling in cardiomyocytes |
7 |
Choline metabolism in cancer |
7 |
Rap1 signaling pathway |
7 |
FoxO signaling pathway |
7 |
Estrogen signaling pathway |
7 |
mRNA surveillance pathway |
7 |
Oxytocin signaling pathway |
7 |
MAPK signaling pathway |
7 |
Neurotrophin signaling pathway |
7 |
Sphingolipid metabolism |
7 |
Insulin signaling pathway |
7 |
HTLV-I infection |
7 |
Tight junction |
7 |
Thyroid hormone synthesis |
7 |
Apoptosis |
7 |
Arrhythmogenic right ventricular cardiomyopathy (ARVC) |
7 |
ErbB signaling pathway |
6 |
Colorectal cancer |
6 |
Renal cell carcinoma |
6 |
Glioma |
6 |
Chronic myeloid leukemia |
6 |
Pancreatic cancer |
6 |
Axon guidance |
6 |
Prostate cancer |
6 |
Melanoma |
6 |
Non-small cell lung cancer |
6 |
Long-term depression |
6 |
Thyroid hormone signaling pathway |
6 |
Hepatitis B |
6 |
Amyotrophic lateral sclerosis (ALS) |
6 |
HIF-1 signaling pathway |
6 |
Endometrial cancer |
5 |
T cell receptor signaling pathway |
5 |
Cell cycle |
5 |
Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate |
5 |
Acute myeloid leukemia |
4 |
Dorso-ventral axis formation |
4 |
Glycosphingolipid biosynthesis - lacto and neolacto series |
4 |
Thyroid cancer |
4 |
Mucin type O-Glycan biosynthesis |
3 |
Biotin metabolism |
1 |
Table 4. Genes correlated with differentially expressed miRNAs and their role(s) in Wnt signal modulation
Target Gene |
Role in Wnt signal modulation |
ROCK1 |
Downstream mediator of non-canonical wnt signaling, major regulator of cytoskeleton mediates actin polymerization, cell polarity and migration. |
SMAD2 |
Effector molecule of TGFβ signaling pathway, involved in canonical Wnt-TGFβ induced fibrotic processes including myofibroblast transformation and extracellular matrix secretion. |
NFAT5 |
NFAT5 is an osmotic stress responder and a transcription factor downstream of non-canonical wnt pathway, regulates cell fate and migration. However NFAT can also inhibit canonical wnt signaling. |
PRKCB |
Wnt pathway regulator, apoptosis inducer. |
TBL1XR1 |
Regulates Wnt-βcatenin mediated transcription as part of a co-repressor complex. |
2021 Copyright OAT. All rights reserv
No specific patterns in the demographic and clinical information and miRNA expression were observed in this small sample.
Discussion
This study highlights the unique miRNA differential profile in the AH of PCG patients after comparison with congenital cataract. miRNA profiling have been previously described in AH [8] and in exosomes isolated from AH [9] in small number of cataract in adult patients. Certain miRNAs including miR-486-5p, miR-184, let-7a, miR-125-5p and miR-181a were found to be common to adult cataract AH in the current study and miRNA in exosomes [8]. With the exception of miR-181a, others were among the top 50 abundant miRNAs. However, none of these miRNAs were found to be differentially expressed between AH from congenital glaucoma and cataract patients in this study. The methods used for detection and analysis, the clinical differences in study population and other unknown variables may be responsible for these variations in miRNA profiles in AH. Another factor that may influence differential expression studies related to AH miRNA is the lack of comparison with AH from individuals without ocular disease as it was ethically not possible to obtain samples from healthy eyes.
Tanaka and colleagues [13] have performed differential miRNA profiling of AH from 10 adult-onset glaucoma, 5 cataract, and 5 patients with epiretinal membrane using the 3D-Gene human miRNA ver. 1.60 chips containing 2019 miRNAs (Toray Industries, Inc., Tokyo, Japan). The study identified 18 differentially expressed miRNAs that were common to both the control and glaucoma groups, of which, 8 were up-regulated and 10 were down-regulated; 3 miRNAs that were significantly up-regulated (~ 2-folds) and were specific only to the glaucoma patients (absent in controls); and 8 miRNAs that were significantly up-regulated (range of 1.6–3 folds) and were detected only in the controls (absent in glaucoma patients). However, none of the differentially expressed miRNAs reported in this study were found to overlap with the 11 differentially expressed miRNAs identified in our study. These discrepancies may arise due to technological and clinical sampling variability. However, it is important to note that the glaucoma patients studied by Tanaka et al. included primary open angle, primary angle closure and pseudoexfoliation glaucoma and none had PCG. This suggests that the miRNA profiling may help differentiate glaucoma types and the miRNAs identified in this study may be highly relevant to the pathogenesis of congenital glaucoma if their biological significance is validated by functional studies.
Unlike previous studies investigating differential miRNA profiling in the AH samples we attempted to look at miRNA profiles using pathway analysis. Our initial analysis had identified 11 differentially expressed miRNAs and 63 different pathways, mainly related to cancer, metabolism and signaling, related to those 11 miRNAs. The detection of 63 different potentially involved biological pathways makes it difficult to assess the importance of these pathways to PCG in particular. However, further analysis using "pathway intersection" tool identified "Wnt signaling pathway" [15]. Wnt signaling was first identified for its role in carcinogenesis, but has since been recognized for its function in embryonic development [16].
Genes related to canonical and non-canonical Wnt pathways are expressed in the human trabecular meshwork (TM) cells and the investigators have shown that all three Wnt signaling pathways are operative in the TM system [17]. Additionally, studies using perfused organ culture eyes linked the Wnt signaling pathways to elevated intraocular pressure (IOP) in this model. The investigators discovered a novel role for secreted frizzled-related protein-1 (sFRP-1), an antagonist of Wnt signaling, in regulating IOP and demonstrated overexpression of sFRP1 in human glaucomatous TM cells and that restoring Wnt signaling in the TM may be a novel disease intervention strategy for treating glaucoma [18]. Additionally, Wnt signaling pathway has a complex relation with the extracellular matrix (ECM). It intersects with TGF-beta pathway to regulate ECM gene expression in multiple cell types and also plays a role in ECM assembly [19]. Evidence from open angle glaucoma studies suggest a functional canonical Wnt signaling in TM and a link between Wnt antagonism and increased TM cell stiffness [20]. It is therefore possible that dysregulation of Wnt signaling pathway in PCG may cause ECM abnormalities in the TM. Further in vitro studies as well as molecular studies of TM specimens from patients with PCG are needed to substantiate this hypothesis. Villareal and colleagues demonstrated that miR-29b played a role in the regulation of ECM formation in the TM and that under in vitro conditions, upregulation of miR-29b might suppress ECM formation [21]. In our study, miR-29b was not detected in the AH of PCG or controls. Other potential indirect evidence that Wnt signaling may be involved in congenital glaucoma is from a case report of congenital glaucoma resulting from deletion of the HBP1 gene. The HBP1 gene, a transcriptional repressor participates in the Wnt signaling pathway [22]; and is expressed in retina, cornea and ciliary body [genecards.org database].
All our PCG patients had mutations in the CYP1B1 gene. However, the exact mechanism of how CYP1B1 mutations contribute to PCG pathogenesis is not fully understood. CYP1B1 is expressed in cornea, ciliary body, iris, and retina. However, no expression has been identified in the TM. Higher expression of CYP1B1 in fetal eyes as compared to the adult eye suggests a role in the maturation of ocular tissues [23]. There is no clear link between Wnt signaling and CYP1B1 although emerging data are connecting Wnt signaling with members of the cytochrome P450 family of enzymes in other cell types, including activation of Wnt signaling by CYP1B1 via transcription factor Sp1[24]. However, links in the TM between Wnt and CYP1B1 are as yet not known.
In our study, the 4 of the 5 miRNAs associated with the Wnt signaling pathway were down-regulated. The interaction between up- and down- regulation of miRNAs and mRNA can be variable. In general, up-regulated miRNAs may silence target genes by interfering with translation without reducing the expression levels of the target mRNAs or mRNA degradation can indeed be induced by miRNAs. Furthermore, additional evidence that the expression of miRNAs decreases the abundance of many transcripts carrying potential miRNA target sites [25]. Based on the known functions of miRNAs expression, it is possible that the down-regulation of miRNAs in the AH of PCG patients causes interference in the Wnt signaling pathway in the TM, resulting in IOP elevation.
It was observed that 3 miRNAs of the miR-548 gene family including miR-548aa, miR-548t-3p and miR-548aj-3p were significantly elevated in PCG patients; with the former two miRNAs showing more than 2-fold increased levels. miR-548aa and miR-548t-3p although located on different chromosomes, 8 and 4 respectively, however, have same mature sequence and target genes. miR-548 consists of a large 69 member, poorly conserved primate-specific miRNA gene family located on almost all the chromosomes except 19 and Y [26]. Functional enrichment analysis have shown that this miRNA gene family plays important roles in multiple biological processes and human diseases including, regulation of actin cytoskeleton, Wnt signaling pathway, ubiquitin mediated proteolysis, TGF-beta signaling pathway, various cancers and Alzheimer’s disease [26]. Considering the specific role of Wnt signaling pathway (as discussed above) and TGF-beta signaling pathway in glaucoma pathogenesis [27] it is possible that miR-548aa and miR-548t-3p may have a role to play in congenital glaucoma via one or more of these pathways.
Other miRNA that also needs a specific mention is miR-638 which showed more than 2-fold decrease in the patient group. Pathway analysis indicated that this miRNA significantly affects drug metabolism pathway involving a single cytochrome P450 gene, CYP3A43. Mutations in CYP1B1 gene has been linked to PCG which is a major cause of congenital glaucoma in the kingdom of Saudi Arabia [3]. However, the role for other cytochrome P450 genes cannot be ruled out [28]. A recent study [22] reported a de novo deletion of 7q22.1 involving a small cluster of cytochrome P450 genes including CYP3A4, CYP3A5, CYP3A7, and CYP3A43 in a young female patient with congenital glaucoma highlighting the importance of other CYP genes and the role miR-638 may have in PCG. Members of the subfamily 3A are expressed in human iris, ciliary body and cornea [29]; and interestingly, CYP3A43 has been shown to be differentially expressed in human cornea [30] and lens. The role of miR-638 and its interaction with the cytochrome P50 family of genes needs to be further investigated.
In conclusion, the study highlights changes in the aqueous humor miRNA profile of patients with congenital glaucoma compared to controls and suggests a plausible role for Wnt signaling pathway in PCG. Further functional studies are needed to identify the biological significance of these miRNAs, particularly miR-548aa, miR548t-3p and miR-638, and their contribution to the pathogenesis of PCG.
Acknowledgments
The authors thank the following KKESH Research Department staff, Dr. Rajiv Khandekar for statistical analysis of patient clinical data, Mr. Gaddah Al Asali, Clinical coordinator for coordinating patient recruitment and Mrs. Maha Othman, Technical Research Associate, for their efforts during the study. Drs Khaled Abu-Amero and Altaf Kondkar are supported by the Glaucoma Research Chair at Dept. of Opthalmology, College of Medicine, King Saud University.
Authors’ contributions
A – research concept and design; B – collection and/or assembly of data; C – data analysis and interpretation; D – writing the article; E – critical revision of the article; F – final approval of article.
Ethics approval and consent to participate
The study adhered to the tenets of the Declaration of Helsinki and had received approval from the Institutional Review Board and Research Ethics Committee. Written, informed consent was obtained from all participants prior to their inclusion in this study.
References
- Kaur K, Mandal AK, Chakrabarti S (2011) Primary Congenital Glaucoma and the Involvement of CYP1B1. Middle East Afr J Ophthalmol 18: 7-16. [Crossref]
- Abu-Amero KK, Osman EA, Mousa A, Wheeler J, Whigham B, et al. (2011) Screening of CYP1B1 and LTBP2 genes in Saudi families with primary congenital glaucoma: genotype-phenotype correlation. Mol Vis 17: 2911-2919. [Crossref]
- Zhao Y, Sorenson CM, Sheibani N (2015) Cytochrome P450 1B1 and Primary Congenital Glaucoma. J Ophthalmic Vis Res 10: 60-67. [Crossref]
- Ali M, McKibbin M, Booth A, Parry DA, Jain P, et al. (2009) Null mutations in LTBP2 cause primary congenital glaucoma. Am J Hum Genet 84: 664-671. [Crossref]
- Bouhenni RA, Al Shahwan S, Morales J, Wakim BT, Chomyk AM, et al. (2011) Identification of differentially expressed proteins in the aqueous humor of primary congenital glaucoma. Exp Eye Res 92: 67-75. [Crossref]
- Izzotti A, Ceccaroli C, Longobardi MG, Micale RT, Pulliero A, et al. (2015) Molecular Damage in Glaucoma: from Anterior to Posterior Eye Segment. The MicroRNA Role. Microrna 4: 3-17. [Crossref]
- Xu S (2009) microRNA expression in the eyes and their significance in relation to functions. Prog Retin Eye Res 28: 87-116. [Crossref]
- Dunmire JJ, Lagouros E, Bouhenni RA, Jones M, Edward DP (2013) MicroRNA in aqueous humor from patients with cataract. Exp Eye Res 108: 68-71. [Crossref]
- Dismuke WM, Challa P, Navarro I, Stamer WD, Liu Y (2015) Human aqueous humor exosomes. Exp Eye Res 132: 73-77. [Crossref]
- Ragusa M, Caltabiano R, Russo A, Puzzo L, Avitabile T, et al. (2013) MicroRNAs in vitreus humor from patients with ocular diseases. Mol Vis 19: 430-440. [Crossref]
- Wu C, Lin H, Wang Q, Chen W, Luo H, et al. (2012) Discrepant expression of microRNAs in transparent and cataractous human lenses. Invest Ophthalmol Vis Sci 53: 3906-3912. [Crossref]
- Li N, Cui J, Duan X, Chen H, Fan F (2012) Suppression of type I collagen expression by miR-29b via PI3K, Akt, and Sp1 pathway in human Tenon's fibroblasts. Invest Ophthalmol Vis Sci 53: 1670-1678. [Crossref]
- Tanaka Y, Tsuda S, Kunikata H2, Sato J, Kokubun T, et al. (2014) Profiles of extracellular miRNAs in the aqueous humor of glaucoma patients assessed with a microarray system. Sci Rep 4: 5089. [Crossref]
- Edward DP, Alkatan H, Rafiq Q, Eberhart C, Al Mesfer S, et al. (2015) MicroRNA profiling in intraocular medulloepitheliomas. PLoS One 10: e0121706. [Crossref]
- Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, et al. (2012) DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res 40: W498-504. [Crossref]
- Li F, Chong ZZ, Maiese K (2006) Winding through the WNT pathway during cellular development and demise. Histol Histopathol 21: 103-124. [Crossref]
- Shyam R, Shen X, Yue BY, Wentz-Hunter KK (2010) Wnt gene expression in human trabecular meshwork cells. Mol Vis 16: 122-129. [Crossref]
- Wang WH, McNatt LG, Pang IH, Millar JC, Hellberg PE, et al. (2008) Increased expression of the WNT antagonist sFRP-1 in glaucoma elevates intraocular pressure. J Clin Invest 118: 1056-1064. [Crossref]
- Tan J, Tong BD, Wu YJ, Xiong W (2014) MicroRNA-29 mediates TGFbeta1-induced extracellular matrix synthesis by targeting wnt/beta-catenin pathway in human orbital fibroblasts. Int J Clin Exp Pathol 7: 7571-7577. [Crossref]
- Morgan JT, Raghunathan VK, Chang YR, Murphy CJ, Russell P (2015) Wnt inhibition induces persistent increases in intrinsic stiffness of human trabecular meshwork cells. Exp Eye Res 132: 174-178. [Crossref]
- Villarreal G, Jr., Chatterjee A, Oh SS, Oh DJ, Kang MH, et al. (2014) Canonical wnt signaling regulates extracellular matrix expression in the trabecular meshwork. Invest Ophthalmol Vis Sci 55: 7433-7440. [Crossref]
- Martinez-Jacobo L, Cordova-Fletes C, Ortiz-Lopez R, Rivas F, Saucedo-Carrasco C, et al. (2013) Delineation of a de novo 7q21.3q31.1 Deletion by CGH-SNP Arrays in a Girl with Multiple Congenital Anomalies Including Severe Glaucoma. Mol Syndromol 4: 285-291. [Crossref]
- Doshi M, Marcus C, Bejjani BA, Edward DP (2006) Immunolocalization of CYP1B1 in normal, human, fetal and adult eyes. Exp Eye Res 82: 24-32. [Crossref]
- Kwon YJ, Baek HS1, Ye DJ1, Shin S1, Kim D2, et al. (2016) CYP1B1 Enhances Cell Proliferation and Metastasis through Induction of EMT and Activation of Wnt/β-Catenin Signaling via Sp1 Upregulation. PLoS One 11: e0151598. [Crossref]
- Wang YP, Li KB (2009) Correlation of expression profiles between microRNAs and mRNA targets using NCI-60 data. BMC Genomics 10: 218. [Crossref]
- Liang T, Guo L, Liu C (2012) Genome-wide analysis of mir-548 gene family reveals evolutionary and functional implications. J Biomed Biotechnol 2012: 679563. [Crossref]
- Kuchtey J, Kuchtey RW (2014) The microfibril hypothesis of glaucoma: implications for treatment of elevated intraocular pressure. J Ocul Pharmacol Ther 30: 170-180. [Crossref]
- Kennedy KD, AnithaChristy SA, Buie LK, Borrás T (2012) Cystatin a, a potential common link for mutant myocilin causative glaucoma. PLoS One 7: e36301. [Crossref]
- Zhang T, Xiang CD, Gale D, Carreiro S, Wu EY, et al. (2008) Drug transporter and cytochrome P450 mRNA expression in human ocular barriers: implications for ocular drug disposition. Drug Metab Dispos 36: 1300-1307. [Crossref]
- Turner HC, Budak MT, Akinci MA, Wolosin JM (2007) Comparative analysis of human conjunctival and corneal epithelial gene expression with oligonucleotide microarrays. Invest Ophthalmol Vis Sci 48: 2050-2061. [Crossref]

