Risk for carcinogenesis in several generations of residents from areas contaminated with radioactive materials was assessed via gene expression profiling of the p53 gene network by the NanoString technology (United States). In total, 24 genes of the TP53 network showed significant differences between the observation and reference groups of subjects. The most significant differences were observed for five genes: ST13, IER3, BRCA1, LRDD, and MRAS. The study confirmed again that the NanoString technology provides overt advantages in terms of sensitivity and throughput when extended gene networks are examined.
ionizing radiation, children, generations I and II of people exposed to ionizing radiation, NanoString technology, p53 gene network, genome instability, carcinogenesis
The NanoString technology has been developed and patented by Krassen Dimitrov and Dwayne Dunaway [1]. The gist is using fluorescent molecular barcodes to detect gene transcripts via fluorescence microscopy, which is capable of visualizing up to 800 nucleic acids per sample. An advantage of the technology is that enzymatic reactions and dilutions are avoided in measuring the mRNA amounts, and measurements are consequently far more sensitive and rapid. Dimitrov and Dunaway’s experiments have shown that the results are highly reproducible in two replicate assays, yielding a replication correlation coefficient of 0.999, a detection limit between 0.1 and 0.5 fM, and a linear dynamic range of 500-fold. Comparisons have demonstrated that the NanoString technology is more sensitive than DNA microarray-based assays and is far simpler and have a higher throughput than real-time PCR using TaqMan and SYBR Green probes.
The NanoString technology is therefore highly efficient in studying large gene networks. Such studies are important to perform in various polygenic disorders, in particular, those due to exposure to external mutagenic factors. Accordingly, the studies may play a material role in evaluating genetic defects and risk for carcinogenesis in people chronically exposed to radiation. The cancer-related p53 gene network is of particular interest to study.
Mammalian p53 originates from a transcription factor that occurred in ancient multicellular organisms and regulated transcription in response to stress as its primary function. A tumor suppressor role of p53 was probably acquired relatively recently in evolution, as an adaptation of organisms that have prolonged lifespans and accumulate many somatic mutations [2]. The significant role that p53 plays in maintaining genome stability is clearly evident from that fact that mutant p53 is found in approximately half of human tumors. More than 10 million tumor samples have been observed to carry inactivated p53, and about the same number of tumors contained other inactivated proteins [3]. However, tumor suppression is not the only function of p53. The protein performs many functions, regulating cell migration, autophagy, anaerobic and aerobic glycolysis, and other cell metabolic pathways [4-6]. In addition, p53 modulates cell differentiation [7]. A p53 deletion suppresses terminal differentiation of skeletal muscles, many hematopoietic cell lineages, renal epithelial cells, thyrocytes, and oligodendrocytes, as well as axon growth and regeneration [8,9]. While often playing a central crucial role in carcinogenesis, the p53 gene (TP53), as any gene, it is involved in interacting with many other genes and their protein products and, with each of them, forms positive and negative feedbacks and compensatory mechanisms, which coordinate the function of the total TP53 network. It is nearly meaningless to study one gene alone without its network, be it even the most important gene.
To demonstrate that gene expression profiling of the p53 gene network with the NanoString technology (United States) is suitable for evaluating risk for carcinogenesis in various generations of residents from areas contaminated with radioactive material.
To assess higher risk for carcinogenesis, we examined 36 residents from Russian areas contaminated with radioactive material as a result of the Chernobyl meltdown. This observation group included 13 mothers, who were born from 1969 to 1987 and formed generation I, and their children (23 in total, one to three per mother), who were younger than 18 years of age and formed generation II. The criteria for inclusion in the monitoring group were also: the permanent residence of patients in the radionuclide contamination region with a soil contamination level of cesium-137 from 556 kBq/m2 to 1665 kBq/m2 (including for the I generation presence at the time of the accident in the indicated Region). Patient generations I and II did not have radiation-induced diseases, including oncopathology.
A reference group included 12 subjects (6 of generation I and 6 of generation II, similar in age) from areas free from radioactive pollution.
To profile expression of the p53 gene network, RNA was isolated from lymphocytes of fresh peripheral blood samples of the 48 subjects. RNA was extracted from peripheral blood with PureLink® Total RNA Blood Kit. Concentration and quality of samples were detected by Implen NanoPhotometer Pearl and gel-electrophoresis.
Samples were run on the nCounter Analysis System (NanoString Technologies), according to the manufacturer’s protocol in a total of 4 runs. Each sample contained at least 100 ng of RNA. All hybridization reactions were 24-26 hours long; all PrepStation run were held on high sensitivity mode and all counts were gathered by scanning on MAX mode for 555 fields of view per sample.
No nCounter samples were flagged by nSolverAnalysisSoftware (NanoString Technologies) for quality control. nCounter data was normalized with inner nCounter-provided positive controls on the basis of RNA yields in each sample. No normalization factors were outside the NanoString-recommended range of 0.3-3. Thresholds to identify expressed genes from background noise were then calculated as the median of the maxima of the negative control probe measurements for each cell type.
We did not confirm our data from nCounter with different methods. Up to date there are numerous papers, that show absolute advantage of Nanostring technology over PCR or microarrays [10,11].
The resulting gene expression data were normalized and statistically analyzed. The analysis was performed using the NCSS 11 statistical package. The results for each gene and sample were checked for normal distribution using the Shapiro-Wilk test. If the distribution with significance level p <0.05 was considered normal, we used the Student's t-test for comparison. If there were not enough evidence for considering the distribution to be normal, we used the nonparametric Mann-Whitney test for comparison.
The p53 gene network includes a huge number of genes, and their list is continuously increasing. To design the panel for the network, we selected the 102 genes most tightly associated with TP53 at the molecular and functional levels according to published data. Six housekeeping genes were chosen as controls. The genes are summarized in the Extended Data Table 1.
Expression profiling of the p53 gene network in generations I and II of residents from radioactive areas was carried out with a digital nCounter Analysis System (NanoString Technologies, United States).
The resulting gene expression data were normalized and statistically analyzed. The analysis was performed using the NCSS 11 statistical package. Data of the two groups were compared by the Mann–Whitney nonparametric test.
The TP53 expression level did not significantly differ between the observation and reference groups. However, expression profiling revealed 24 genes that showed statistically significant differences between the two groups (Figure 1).
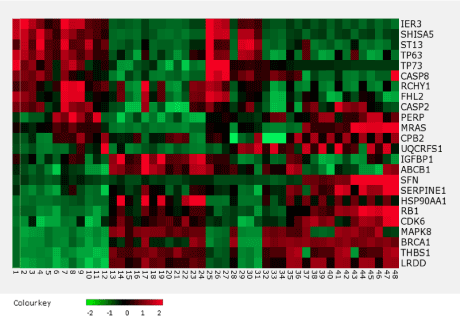
Figure 1. The heatmap of the p53 gene network that showed significant (p < 0.05) upregulation or downregulation.
The most significant differences were observed for five genes: ST13, IER3, BRCA1, LRDD, and MRAS (Figure 2).
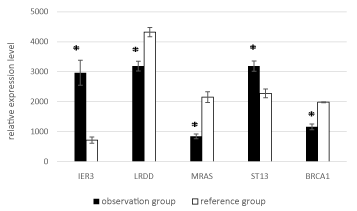
Figure 2. Mean expression levels of ST13, IER3, BRCA1, LRDD, MRAS in the observation and reference groups. Hereafter, the whiskers show the standard error of the mean (SEM)); * - for significant (p < 0.05) differences between groups.
Stratified by generation, the expression levels of all five genes significantly differed significantly between the two groups in both generation I and generation II (Figures 3 and 4).
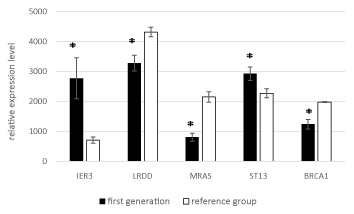
Figure 3. Mean expression levels of the five genes in generation I to the reference group.
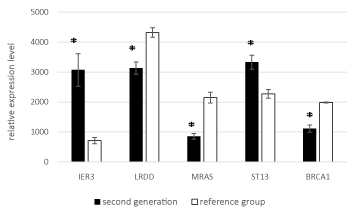
Figure 4. Mean expression levels of the five genes in generation II to the reference group.
The two generations did not significantly differ in gene expression levels (Figure 5).
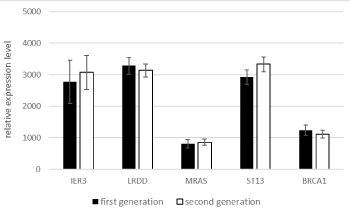
Figure 5. Mean expression levels of the five genes in generations I and II of the observation group.
2021 Copyright OAT. All rights reserv
Thus, our results show that TP53 expression in the two generations of residents from radioactive areas does not differ from that in the reference group. However, the analysis of the p53 gene network demonstrated that significant changes (upregulation or downregulation) occur in expression of several functionally important genes and are possibly transmitted from one generation to the next one.
The analysis using the NanoString technology revealed 24 genes with significant changes in expression in the p53 gene network. Five genes showed the most substantial changes.
A significant increase in the level of gene expression:
IER3 is expressed to a high level in many tumor tissues. Higher levels of its protein product may increase risk for tumorigenesis. Its role in regulating apoptosis is a matter of active discussion. IER3 expression is tightly associated with mutations of TP53, Ki-67, and EGFR, leading to a greater tumor size and a more advanced tumor stage [12].
ST13 which inhibits tumor growth in various cancer; ST13 downregulation may increase risk for gastric and colorectal carcinomas [13]. The gene codes for an adaptor protein that interacts with the heat shock proteins and is involved in glucocorticoid receptor assembly via many molecular chaperones. The expression of this gene in the observation group is increased relative to the values in the reference group, which may be the result of the response of the organism to the long-existing high risk of developing a tumor or radiation-induced activation of carcinogenesis processes.
The expression level of the following genes is significantly lowered:
BRCA1 downregulation may increase risk for breast and ovarian cancers. The gene codes for a phosphoprotein that maintains genome stability and acts as a tumor suppressor. BRCA1 mutations are found in approximately 40% of inherited breast cancer cases. Downregulation or lack of BRCA1 expression is observed in the majority of breast cancer cases [14].
LRDD Downregulation of the gene may decrease apoptotic activity in response to genotoxic agents. The protein product is also known as a p53-induced protein with a death domain. The protein can interact with other death domain-possessing proteins (such as FADD or MADD) in signaling cascades and leads to spontaneous activation of caspase 2 [15,16].
MRAS Downregulation of the gene also may decrease apoptotic activity. The gene codes for a protein of the Ras family (small GTPases). The product is a membrane protein and acts as a transducer in many processes, including cell growth and differentiation. Ras deregulation is associated with many cancers. The protein plays an important role in the TNF-α and MAP kinase signaling pathways [provided by RefSeq, Nov 2011]
Thus, our results show that TP53 expression in the two generations of residents from radioactive areas does not differ from that in the reference group. However, the analysis of the p53 gene network demonstrated that significant changes (upregulation or downregulation) occur in expression of several functionally important genes and are possibly transmitted from one generation to the next one.
Changes in expression of four out of the five genes in question relative to the reference group suggest higher risk of carcinogenesis for residents of areas contaminated with radioactive materials. The other genes need further investigation in larger samples of subjects and at the DNA level with the use of high-throughput sequencing.
A correlation analysis of the results is now in progress; its results will soon be published.
Our pilot study with the NanoString technology makes it possible to assume a transgenerational transmission for specific consequences of radiation exposure and, therefore, higher risk of carcinogenesis.
Our study testifies again that the NanoString technology in advantageous in terms of both sensitivity and throughput for examining large gene networks.
Thus, an important and pressing task now is to continue the study with more gene networks involved and a more detailed analysis of their genes.
This work was supported by the Russian Foundation for Basic Research (project № 15-54-50006).
View supplementary data
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, et al. (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26: 317-325. [Crossref]
- Amaral JD, Xavier JM, Steer CJ, Rodrigues CM (2010) The role of p53 in apoptosis. Discov Med 9: 145-152. [Crossref]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP (2009) Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer 9: 862-873. [Crossref]
- Armata HL, Golebiowski D, Jung DY, Ko HJ, Kim JK, et al. (2010) Requirement of the ATM/p53 tumor suppressor pathway for glucose homeostasis. Mol Cell Biol 30: 5787-5794. [Crossref]
- Aylon Y, Yabuta N, Besserglick H, Buganim Y, Rotter V, et al. (2009) Silencing of the Lats2 tumor suppressor overrides a p53-dependent oncogenic stress checkpoint and enables mutant H-Ras-driven cell transformation. Oncogene 28: 4469-4479. [Crossref]
- Olovnikov IA1, Kravchenko JE, Chumakov PM (2009) Homeostatic functions of the p53 tumor suppressor: regulation of energy metabolism and antioxidant defense. Semin Cancer Biol 19: 32-41. [Crossref]
- Molchadsky A1, Shats I, Goldfinger N, Pevsner-Fischer M, Olson M, et al. (2008) p53 plays a role in mesenchymal differentiation programs, in a cell fate dependent manner. PLoS One 3: e3707. [Crossref]
- Almog N, Li R, Peled A, Schwartz D, Wolkowicz R, et al. (1997) The murine C'-terminally alternatively spliced form of p53 induces atten-uated apoptosis in myeloid cells. Mol Cell Biol 17: 713-722. [Crossref]
- Molchadsky A1, Rivlin N, Brosh R, Rotter V, Sarig R, et al. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis 31: 1501-1508. [Crossref]
- Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, et al. (2015) Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. Cancer Res 75: 2587-2593. [Crossref]
- Bentley-Hewitt KL, Hedderley DI, Monro J, Martell S, Smith H, et al. (2016) Comparison of quantitative real-time polymerase chain reaction with NanoString® methodology using adipose and liver tissues from rats fed seaweed. N Biotechnol 33: 380-386. [Crossref]
- Liu Z, Wang XM, Jia TF, Zhai Y, Sun LY, et al. (2015) Expression of IER3 in primary hepatocarcinoma: correlation with clinico-pathological parameters. Asian Pac J Cancer Prev 16: 679-682. [Crossref]
- Wang LB, Zheng S, Zhang SZ, Peng JP, Ye F, et al. (2005) Expression of ST13 in colorectal cancer and adjacent normal tissues. World J Gastroenterol 11: 336-339. [Crossref]
- Hedau S, Batra M, Singh UR, Bharti AC, Ray A, et al. (2015) Expression of BRCA1 and BRCA2 proteins and their correlation with clinical staging in breast cancer. J Cancer Res Ther 11: 158-163. [Crossref]
- Lin Y, Ma W, Benchimol S (2000) Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet 26: 122-127. [Crossref]
- Tinel A, Tschopp J (2004) The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304: 843-846. [Crossref]





