Introduction :The Myocardial Perfusion Imaging (MPI) is a non-invasive image test that allows the assessment of perfusion, function and viability of the Left Ventricle (LV). The quantitative parameters obtained post-reconstruction requires an accurate segmentation of the LV. ImageJ is an open-source software that provides segmentation techniques that may contribute to the segmentation of the LV in the MPI. The purpose of this study was to study the influence of the different segmentation methods provided by ImageJ, in MPI, depending on the administered activity.
Material and methods : We carried out an experimental research with 4 MPI studies simulated with 275, 385, 500 and 750 Bq/voxel in the myocardium, whose short-axis (SA) slices were segmented with ImageJ by the threshold default, OTSU and k-means Plugin Toolkit methods (k=2, k=3). To analyze the most appropriate segmentation method, the signal-to-noise ratio (SNR) for each short-axis (SA) slice was calculated, in accordance with the slices obtained from the software Quantitative Perfusion Single Photon Emission Computed Tomography® (QPS®) and by manual segmentation using ImageJ. To analyze the SNR with ImageJ and QPS® segmentation methods in the same simulated study, and to compare with the same segmentation method in different simulated studies, the Friedman and Kruskal-Wallis tests were applied.
Results and discussion: The method k-means with k=3 is the most suitable method for the segmentation of the LV, regardless of the administered activity.
Conclusion :This study may contribute to the clinical implementation of open-source based segmentation methods of the LV in MPI, according to the activity in the myocardium.
imageJ; myocardial perfusion imaging; segmentation; signal-to-noise ratio
Segmentation is an image processing technique which aims to separate Regions of Interest (ROI) of a given image, which differ among themselves according to properties of discontinuity, such as color scale and brightness. It can be divided into manual, semi-automated and fully-automated methods [1-10]. The myocardial perfusion imaging (MPI) is a Nuclear Medicine test that aims to assess perfusion, function and viability of the Left Ventricle (LV), based on the analysis of quantitative parameters of perfusion and of function, obtained by validated softwares, such as Quantitative Gated/Perfusion Single Photon Emission Computed Tomography® (QGS®/QPS®) [11-13].
This exam requires the administration of a radiopharmaceutical, usually 99mTc-Tetrofosmin or 99mTc-Sestamibi, and a post-acquisition Single-Photon Emission Computerized Tomography (SPECT) [11-13]. From the reconstructed tomographic study, the segmentation of the LV is performed in the Short-Axis slices (SA) in order to extract the quantitative parameters. Therefore, the correct segmentation of LV is strictly necessary [14,15]. Manual segmentation is often not applied to the LV in MPI mainly due to its limited reproducibility, it should be preferred the application of a robust, convergent and highly accurate semi-automated or automated method [16,17]. However, an ideal segmentation method has not been proclaimed yet, due to controversies surrounding the administered activities in MPI (444-1480 MBq) [9,16-22].
Nowadays the potential qualities of open-source software’s can be applied for the segmentation of LV to serve the purposes of Nuclear Cardiology (NC) [23]. ImageJ is a public, open-source software of image processing and analysis, completely free, which includes useful tools of segmentation techniques, that can be applied to NC [24,25]. The purpose of this research was to study the influence of different segmentation methods provided by ImageJ (threshold default and OTSU originally inserted in the ImageJ software, and k-means plugin toolkit), in MPI, liable on the simulated administered activity (275, 385, 500 and 750 Bq/voxel in the myocardium).
Obtaining SA slices from the LV
We carried out an experimental research in 4 GATE (Geant4 Application for Tomography Emission) Monte Carlo simulations of MPI Gated-SPECT studies using the NCAT (Non-Uniform Rational Basis Spline based Cardiac-Torso) phantom [14,26]. The radioactive source used in the studies was the 99mTc-Tetrofosmin [27]. Simulations of MPI studies were simulated with 275, 385, 500 and 750 Bq/voxel in the myocardium, which correspond, respectively, to the total administration of 250, 350, 450 and 680 MBq in the human body, with a constant SPECT projection time of 15 seconds [26]. Using a medical imaging workstation from GE Medical Systems, Xeleris Functional Imaging Workstation (Xeleris® TM 1.0628), the studies were reconstructed by filtered backprojection, with butterworth prefilter, order 5 and cutoff frequency of 0.52 cycles.centimeter-1 (cycles.cm-1), without attenuation correction, as recommended by the manufacturer settings in the commercial implementation of QPS®/QGS®. Perfusion SA slices with the thickness of 6 millimeters (mm) were exported into the ImageJ software version 1.8.0_60 for Windows, using the application RadiAnt DICOM Viewer version 3.0.2. [28].
Methodology for evaluation of applied segmentation methods available in ImageJ
The images were converted to 8 bits and segmented by the threshold default, the OTSU method and the k-means (k=2 and k=3) plugin toolkit in ImageJ software. In order to study if there were differences between the non-segmented studies and segmented studies, a manual ROI was drawn for the total of 14 SA slices, radially, from the apex to the base and determined its area (see figure 1). To analyze the most similar segmentation method available on ImageJ relative to the validated software QPS®, the signal-noise ratio (SNR) of each SA slice was calculated according to the SNR._jar plugin, that applies the formula SNR of Gonzalez [29]:
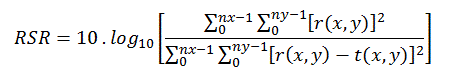
Figure 1. Example of manual ROI design in the LV: in SA slice of one non-segmented study (a) and segmented study by k-means method with k=2 (b).
Where r(x,y) corresponds to the reference image and t(x,y) to the image in which we intend to calculate the SNR; and both images must have the same dimensions [nx,ny]. Specifically in our trial, the r(x,y) is the SA slices segmented with QPS®, and the t(x,y) is the SA slices manually segmented by an operator, using the ImageJ segmentation methods.
The data were analyzed using the statistical software SPSS (Statistical Package for Social Sciences) version 22.0 for Windows. The results were considered significant, with a significance level of 5%. To test the normality of the data, we used the Shapiro-Wilk adjustment test, and since the normality assumptions were not met (p <0.05), we opted for the application of nonparametric statistical techniques. To compare the SNR with different methods in the same study, we used the Friedman test. When statistically significant differences were detected, multiple comparisons for Friedman test was applied. To compare SNR with different simulated studies segmented by the same method, we used the Kruskal-Wallis test. When statistically significant differences were detected, the multiple comparisons for Kruskal-Wallis test was applied.
Table 1 shows the descriptive analysis relative to the areas of the ROI manually drawn in the LV in all SA slices from the apex to the base of the LV of the simulated studies, non-segmented and segmented. Based on the descriptive measures of LV’s ROI area (see table 1), there are evident differences when different segmentation methods are applied in the same study, particularly between non-segmented and segmented studies. However, the results obtained by the threshold default and OTSU segmentation methods were similar in all cases. The study simulated with 385 Bq/voxel in the myocardium showed the lowest mean of the area of the ROI, in every segmentation method, while the study simulated with 275 Bq/voxel in the myocardium was the study which showed the highest mean of the area of the ROI, despite also being the study that showed the highest standard deviation in all segmentation methods.
Table 1: Descriptive analysis of the areas of the ROI drawn in the LV in all SA slices (in mm2).
|
Myocardium voxel activity (Bq/voxel) |
275 |
385 |
500 |
750 |
Minimum (mm2) |
Maximum
(mm2) |
Mean
(mm2) |
Standard
deviation
(mm2) |
Minimum
(mm2) |
Maximum
(mm2) |
Mean
(mm2) |
Standard
deviation
(mm2) |
Minimum
(mm2) |
Maximum
(mm2) |
Mean
(mm2) |
Standard
deviation
(mm2) |
Minimum
(mm2) |
Maximum
(mm2) |
Mean
(mm2) |
Standard
deviation
(mm2) |
N-segm * |
8177.34 |
17879.27 |
15228.12 |
2829.24 |
8131.14 |
15245.89 |
13220.21 |
2243.33 |
9886.73 |
15892.69 |
13874.12 |
1994.66 |
8870.34 |
17555.87 |
15590.61 |
2497.18 |
Threshold
default |
1663.19 |
13813.70 |
9076.46 |
3447.87 |
3233.98 |
9701.93 |
7338.64 |
1955.89 |
4804.77 |
10579.72 |
7519.88 |
1879.78 |
3973.17 |
10949.32 |
7665.59 |
2171.59 |
OTSU |
1663.19 |
12935.91 |
8401.23 |
3177.93 |
3233.98 |
10025.33 |
7271.12 |
1976.94 |
4804.77 |
10579.72 |
7466.58 |
1859.36 |
3973.17 |
11365.12 |
7633.61 |
2195.10 |
k=2 |
52.00 |
251.00 |
166.69 |
54.76 |
70.00 |
202.00 |
154.23 |
39.95 |
104.00 |
229.00 |
162.69 |
40.66 |
86.00 |
237.00 |
163.00 |
45.51 |
k=3 |
136.00 |
330.00 |
250.46 |
69.41 |
115.00 |
267.00 |
212.23 |
50.87 |
177.00 |
267.00 |
226.15 |
33.02 |
136.00 |
325.00 |
244.85 |
59.73 |
* N-segm, non-segmented.
We compared the SNR with QPS and manual SNR for each simulated study non-segmented and segmented by different methods. Multiple comparison for Friedman test allowed us to identify the segmentation methods that differed significantly not only among themselves but also with non-segmented studies (see table 2). A look at table 2 lets us conclude that, taking into consideration the SNR with QPS, every segmentation method applied differs from non-segmented studies except for the k-means with k=3 segmentation method, and that statistically significant differences between the threshold default and k-means with k=3 segmentation methods were identified in the studies simulated with 275 and 385 Bq/voxel in the myocardium.
Table 2: Comparison of the SNR with QPS and manual SNR with different segmentation methods applied in the same study and with non-segmented studies, using the multiple comparison for Friedman test.
Myocardium voxel activity (Bq/voxel) |
Between |
SNR with QPS |
Manual SNR |
Test statistic |
Multiple comparison (p) |
Test statistic |
Multiple comparison (p) |
275 |
Threshold default |
N-segm * |
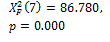
|
0.000 † |
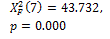
|
0.000 † |
OTSU |
1.000 |
1.000 |
k=2 |
0.699 |
1.000 |
k=3 |
0.029 † |
1.000 |
OTSU |
N-segm * |
0.000 † |
0.006 † |
k=2 |
1.000 |
1.000 |
k=3 |
0.097 |
1.000 |
k=2 |
N-segm * |
0.005 † |
0.000 † |
k=3 |
1.000 |
1.000 |
k=3 |
N-segm * |
0.182 |
0.000 † |
385 |
Threshold default |
N-segm * |
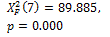
|
0.000 † |
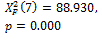
|
0.000 † |
OTSU |
1.000 |
1.000 |
k=2 |
0.567 |
0.567 |
k=3 |
0.022 † |
0.012 † |
OTSU |
N-segm * |
0.000 † |
0.000 † |
k=2 |
1.000 |
1.000 |
k=3 |
0.075 |
0.044 † |
k=2 |
N-segm * |
0.001 † |
0.001 † |
k=3 |
1.000 |
1.000 |
k=3 |
N-segm * |
0.050 |
0.085 |
500 |
Threshold default |
N-segm * |
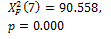
|
0.000 † |
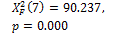
|
0.000 † |
OTSU |
1.000 |
1.000 |
k=2 |
1.000 |
1.000 |
k=3 |
0.066 |
0.050 |
OTSU |
N-segm * |
0.000 † |
0.000 † |
k=2 |
1.000 |
1.000 |
k=3 |
0.066 |
0.050 |
k=2 |
N-segm * |
0.001 † |
0.001 † |
k=3 |
1.000 |
1.000 |
k=3 |
N-segm * |
0.050 |
0.066 |
750 |
Threshold default |
N-segm * |
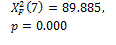
|
0.000 † |
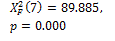
|
0.000 † |
OTSU |
1.000 |
1.000 |
k=2 |
1.000 |
1.000 |
k=3 |
0.075 |
0.075 |
OTSU |
N-segm * |
0.000 † |
0.000 † |
k=2 |
1.000 |
1.000 |
k=3 |
0.075 |
0.075 |
k=2 |
N-segm * |
0.001 † |
0.001 † |
k=3 |
1.000 |
1.000 |
k=3 |
N-segm * |
0.050 |
0.050 |
* N-segm, non-segmented.
† Statistically significant differences for p<0,05.
Taking into consideration the manual SNR, in the study simulated with 275 Bq/voxel in the myocardium every segmentation method applied differed from the non-segmented study; in the remaining simulated studies, it was found that every segmentation method, except for the k-means with k=3 method, showed statistically significant differences when compared with non-segmented studies. In the study simulated with 385 Bq/voxel in the myocardium there was a statistically significant difference between the threshold default and k-means with k=3 segmentation methods and also between the OTSU and k-means with k=3 segmentation methods. Regarding the SNR with QPS, statistically significant differences between the OTSU and k-means with k=3 were never identified.
M. Hosbtalab et al. [16] defend that threshold segmentation is not suitable for the segmentation of the LV, while OTSU segmentation is more accurate and therefore suitable. However, no significant differences were observed in the studies simulated using ImageJ’s segmentation methods. For each segmentation method, we compared the SNR with QPS and manual SNR between the studies simulated with different activities in the myocardium. Multiple comparisons for Kruskal-Wallis test allowed us to identify for each segmentation method the respective simulated studies that showcased statistically significant differences (see table 3). Table 3 shows that there is a greater number of significant differences between the simulated studies segmented by the same method taking into account the manual SNR rather than the SNR with QPS; most likely due to intra-operator variance in the manual segmentation of SA slices of the different simulated studies used as reference images for calculating the manual SNR [3,30,31].
Table 3: Comparison of the SNR with QPS and manual SNR with the segmented studies by the same method, using the multiple comparison for Kruskal-Wallis test.
Segmentation methods |
Between |
SNR with QPS |
Manual SNR |
Myocardium voxel activity (Bq/voxel) |
Myocardium voxel activity (Bq/voxel) |
Test statistic |
Multiple comparison (p) |
Test statistic |
Multiple comparison (p) |
Threshold default |
275 |
385 |
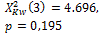
|
|
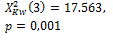
|
0.004 * |
500 |
0.002 * |
750 |
0.007 * |
385 |
500 |
1.000 |
750 |
1.000 |
500 |
750 |
1.000 |
OTSU |
275 |
385 |
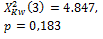
|
|
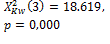
|
0.004 * |
500 |
0.001 * |
750 |
0.004 * |
385 |
500 |
1.000 |
750 |
1.000 |
500 |
750 |
1.000 |
k=2 |
275 |
385 |
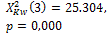
|
0.006 * |
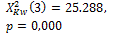
|
0.005 * |
500 |
1.000 |
0.000 * |
750 |
0.000 * |
0.000 * |
385 |
500 |
0.062 |
1.000 |
750 |
1.000 |
1.000 |
500 |
750 |
0.002 * |
1.000 |
k=3 |
275 |
385 |
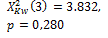
|
|
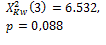
|
|
500 |
750 |
385 |
500 |
750 |
500 |
750 |
* Statistically significant differences for p<0,05.
Taking into account the SNR with QPS, statistically significant differences were only identified in the k-means with k=2 method, between the studies simulated with 275 and 385 Bq/voxel in the myocardium, with 275 and 750 Bq/voxel in the myocardium and between the studies simulated with 500 and 750 Bq/voxel in the myocardium.
Regarding to the manual SNR in the threshold default, OTSU and k-means with k=2 segmentation methods, the study simulated with 275 Bq/voxel in the myocardium differed significantly from the other simulated studies.
Figure 2 shows the box plots of distribution of SNR with QPS and manual SNR, for each segmentation method, applied in the 4 simulated studies. Considering the formula of calculation of SNR’s Gonzalez, the higher the value of SNR the better the quality of the images. When the logarithm of the function varies between 0 and 1 the value of SNR is negative, and on the other hand, when it is more than 1, the value of SNR is positive. Accordingly, in regard to the distribution of SNR with QPS and manual SNR evidenced in figure 2, it can be verified that the threshold default and OTSU segmentation methods always presented negative values, unlike the k-means methods. It was also observed that all segmentation methods damaged the quality of the SA slices of all simulated studies. However, irrespectively of the myocardium voxel activity of the simulated studies, when segmented by any of the k-means methods, they present a better distribution of SNR with QPS and of manual SNR, particularly when k-means with k=3 segmentation is applied. This last observation is in agreement with the view held by R. Alves [9].
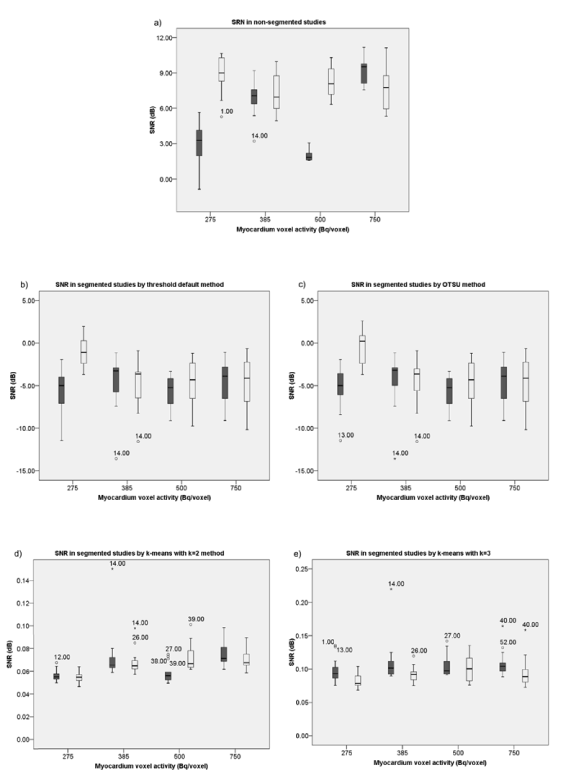
Figure 2. Distribution of SNR with QPS ( ) and manual SNR ( ) in the studies: non-segmented (a), segmented by default threshold (b), OTSU (c), k-means with k=2 (d) and k=3 (e).
It was observed that when the threshold default and OTSU methods were applied, they yielded better distributions of SNR with QPS in the simulated study with 385 Bq/voxel in the myocardium, and for the manual SNR, its best distributions were observed in the simulated study with 275 Bq/voxel in the myocardium. When the k-means with k=2 segmentation method was applied it obtained a better distribution of SNR with QPS and manual SNR in the simulated study with 750 Bq/voxel in the myocardium. When the k-means with k=3 segmentation method was applied, a better distribution of SNR with QPS and of manual SNR were obtained in the study simulated with 500 Bq/voxel in the myocardium.
According to the study by S. Underwood et al. [18], it would be expected that with the increase of activity in the myocardium, a gradual increase of the SNR existed for all segmentation methods applied. However, this was not observed in any of the methods nor in non-segmentation studies, probably because S. Underwood et al. focused their study to real MPI studies, and the simulated studies disregard the background radiation and other errors associated, like the physiology, pathophysiology and heterogeneity of human beings, in their anatomy and in the biodistribution of radiopharmaceuticals.
The segmentation of the LV in QPS® software can only involve the use of threshold segmentation, and only if it is not successful, can a cluster segmentation method be used as well; according to the study performed on ImageJ, the k-means segmentation (with k=2 or k=3, but with better results with k=3) presented a lower SNR with QPS and manual SNR in comparison with the threshold default segmentation method, and because of that, it is considered more efficient in the segmentation of the LV.
Therefore, in regards to this study, a greater SNR is obtained in studies whose total administered activity is 450 MBq (corresponding to 500 Bq/voxel in the myocardium), which is in agreement with the recommendations of the EANM guidelines regarding the activity administration on the first phase of the one-day protocol of MPI study [11-12].
In this study, we conclude that the most suitable method for the segmentation of the LV, using the ImageJ software, is the k-means with k=3 method, regardless of the simulated administered activity. This study comes off as innovative in the sense that may contribute to the clinical implementation of open-source based segmentation methods of the LV in MPI, according to the activity in the myocardium.
However, we suggest the application of new lines of research, involving real clinical MPI studies.
2021 Copyright OAT. All rights reserv
- Boudraa AO, Zaidi H (2006) Image Segmentation Techniques in Nuclear Medicine Imaging. In: Zaidi, H., ed. 2006. Quantitative analysis in Nuclear Medicine Imaging. Springer US, pp.308–357. Ch.10.
- Torres SA (2012) Segmentação de Imagens Médicas Visando a Construção de Modelos Médicos. DEd. Instituto Politécnico de Bragança-Escola Superior de Tecnologia e Gestão.
- Rodrigues, G. and Ventura, S.R., 2010. Técnicas de Processamento de Imagem em Cardio-TC e Cardio-RM. TDTOnline Magazine. 2, pp.5-8.
- Facon J, Limiarização bimodal de OTSU. [pdf] Paraná: Universidade Católica do Paraná.
- Otsu N (1979) A Threshold Selection Method from Gray-Level Histograms. Institute of Electrical and Electronics Engineers Xplore 20.
- Torok L, Método de Otsu. [pdf] Rio de Janeiro: Universidade Federal Fluminense.
- Assunção R (2013) Segmentação de Imagens Dentárias por Binarização, agrupamento de contornos ativos. DEd. Instituto Politécnico de Bragança-Escola Superior de Tecnologia e Gestão.
- Azevedo PA (2010) Clustering. [PowerPoint] Minho: Universidade do Minho.
- Alves R (2014) Analysis of gated myocardial perfusion spect images using computational image registration techniques. DEd. Faculdade de Engenharia da Universidade do Porto.
- Faria D, Tavares J, Costa DC (2011) Segmentação, Reconstrução e Quantificação 3D – Aplicação em Imagem Funcional e Metabólica. In: XIII Congresso Nacional de Medicina Nuclear. Cascais
- Ryder H, Testanera G, Jerónimo VV, Vidovič B eds. 2014. Myocardial Perfusion Imaging. Austria: European Association of Nuclear Medicine
- Hesse B, Tägil, K., Cuocolo A. et al., 2005. EANM/ESC procedural guidelines for myocardial perfusion imaging in nuclear cardiology. European Journal of Nuclear Medicine and Molecular Imaging 32.
- Verberne H J, Acampa W, Anagnostopoulos C (2015) EANM procedural guidelines for radionuclide myocardial perfusion imaging with SPECT and SPECT/CT: 2015 reversion. European Journal of Nuclear Medicine and Molecular Imaging. 42:12.
- Vieira, L (2010) Contributo para a optimização dos estudos de perfusão miocárdica utilizando imagens de medicina nuclear sincronizadas com o electrocardiograma. Ph. D., Universidade de Lisboa.
- Germano G (2006) Artificial Intelligence Techniques in Nuclear Cardiology. Cardiac Imaging 36.
- Hosntalab M, Babapour-Mofrad F, Monshizadeh N, Amoui M (2012) Automatic left ventricle segmentation in volumetric SPECT data set by variational level set. International Journal of Computer Assisted Radiology and Surgery 7:6.
- Germano G (1995) Automatic Quantification of Ejection Fraction from Gated Myocardial Perfusion SPECT. Journal Nuclear Medicine 36:11.
- Underwood SR, Robinson CN, Aswegen A van (2008) The relationship between administered radiopharmaceutical activity in myocardial perfusion scintigraphy and imaging outcome. European Journal of Nuclear Medicine and Molecular Imaging 35:2.
- Choi S (2007) Segmentation of the left ventricle in myocardial perfusion SPECT using variational level set formulation. In: Institute of Electrical and Electronics Engineers Nuclear Science Symposium Conference Record. Honolulu, 26 October–3 November 2007, 4, pp.3060–3064 .
- Liu YH, Sinusas AJ, Khaimov D (2005) New hybrid count- and geometry-based method for quantification of left ventricular volumes and ejection fraction from ECG-gated SPECT : methodology and validation. Journal of Nuclear Cardiology 12.
- Tan WH, Besar R (2009) Segmentation of the Left Ventricle in Myocardial Perfusion SPECT Using Active Shape Model. In Zaman, H.B., Robinson, P., Petrou, M. et al., eds. 2009. Visual Informatics: Bridging Research and Practice. Malasya: Springer. pp.38–49.
- Germano G, Kavanagh PB, Waechter P (2000) A new algorithm for the quantitation of myocardial perfusion SPECT. I: technical principles and reproducibility. Journal of Nuclear Medicine 41: 4.
- Nagy P (2007) Open source in imaging informatics. Journal of Digital Imaging. 20:1.
- Ferreira T, Rasband W (2012) ImageJ User Guide.
- Rasband W, Eliceiri K, Schneider CA (2012 ) NIH Image to ImageJ : 25 years of image analysis. Nature Methods 9.
- Vieira L, Costa DC, Almeida P (2015) The influence of number of counts in the myocardium in the determination of reproducible functional parameters in gated-SPECT studies simulated with GATE. Revista Española de Medicina Nuclear e Imagem Molecular 34-6.
- Higley B, Smith FW, Smith, T (1993) Technetium-99m-1,2-bis[bis(2-ethoxyethyl) phosphino] ethane: human biodistribution, dosimetry and safety of a new myocardial perfusion imaging agent. Journal of Nuclear Medicine 34:1.
- Medixant (2016) RadiAnt DICOM Viewer.
- Sage D (2013) SNR_.jar. [computer program] École Polytechnique Fédérale de Lausanne.
- Reimão AS, Pereira J, Nobre M (2015) Influência da experiência profissional e da função visual do operador no processamento semiautomático da cintigrafia de perfusão do miocárdio: variabilidade intra e interoperador na determinação dos parâmetros quantitivos. Revista Saúde & Tecnologia 14.
- Foster B, Bagci U, Mansoor A (2014) A review on segmentation of positron emission tomography images. Computers in Biology and Medicine 50.


