Infections with Respiratory syncytial virus may result in mild to severe illness with lower respiratory involvement leading to bronchiolitis and pneumonia which are the main cause of respiratory symptoms. The aim of the study was to detect of RSVgenotypes in the hospitalized children with bronchiolitis in Khartoum hospitals .The study included 92 subjects, 21 samples were positive by using real time PCR and for RSV-A and 2 samples werepositive for RSV-B by using nested PCR .The percentage of male to female patient was 52: 48, Children less than 2 years were found to be more susceptible to RSVs infections and RSV A was the predominant genotype and was found more commonly in male than female.
RSV, bronchiolitis, real-time PCR, Sudan
Acute lower respiratory tract infection (ALRI) is a severe disease that leads to morbidity and mortality in infants and children worldwide. One of the most common causes of ALRI is human respiratory syncytial virus (HRSV) [1,2]. Respiratory syncytial virus (RSV) of the family Paramyxoviridae, is a non-segmented, negative strand RNA virus that expresses 11 proteins [3], with an RNA genome of approximately 15.2 Kb. The surface proteins of the virus include the G (glycoprotein), F (fusion protein) and SH (small hydrophobic protein). The G protein is type II glycoprotein that is involved in attachment of virion to the host cell [4-6], RSV is the most common cause of bronchiolitis and pneumonia in children younger than 1 year [7]. Infants born preterm, and children with chronic lung disease (CLD), congenital heart disease (CHD), or other high-risk medical conditions have a higher risk of developing more severe RSV infections [8].
RSV has been classified into two antigenic subgroups; A and B, initially on the basis of the reactivity of the virus to monoclonal antibodies directed against the attachment glycoprotein (G protein) [9], and currently through genetic analyses [10].
In developing countries, there are a few population based estimates of the incidence of RSV disease. Data from a small number of studies has placed the estimated incidence of RSV-associated lower respiratory infections (LRI) at around 97 to 180 episodes per 1000 child / year which is a crude estimate, at best. However, these estimated rates for developing countries are 2.6 to 4.8 times the rate of RSV associated LRI seen in the USA [11]. Laboratory methods available for the detection of RSV include, virus isolation in cell culture, detection of viral antigens by direct or indirect immunofluorescent (IF) staining (DFA/IFA) or by enzyme-linked immunosorbent assays (ELISA) and the detection of viral nucleic acids by amplification assays [12], mainly reverse transcription polymerase chain reaction (RT-PCR) and real time polymerase chain reaction which represent rapid and sensitive methods for detection of RSV compared to other techniques [13].
Ethical approval
The study has been approved by the local ethics committee of Al-neelain University. All parents of participants in the study were given a written informed consent considering the aims of the study, Sample and clinical information’s were used anonymously.
Study area
The study was conducted in Khartoum State of Central Sudan, Patients involving children under 5 years of age with respiratory tract infection (RTI, and were divided in to two age groups, at the Mohamed Alamin hamid pediatric hospital in Omdurman were recruited during the period December 2015 to May 2016.
Data collection
Demographic data of the patients were collected using a structured questionnaire, which included the following criteria: Age, gender, and clinical symptoms (bronchiolitis). The specimens were collected within 1-7 days of the disease onset.
Collection of specimens
A total of 92 nasal swab samples were collected from patients by using sterile nylon swabs (Regular Flocked swab, Cat. No. 520CS01, Copan Diagnostics Inc., Murrieta, Calif, USA) in 1 ml of transport media (UTM-RT, Cat. No. 92562, Copan Diagnostics Inc., Murrieta, Calif, USA). Samples collected were transported on ice at the same day of collection to the Department of Virology, Central Laboratory (Ministry of high education) and stored at −80°C until processed.
RNA extraction
RNA was extracted from nasal swab samples using QIAamp Viral RNA Mini Kit according to manufacturer’s instructions.
Real time Polymerase Chain Reaction
The Real-time RT-PCR was performed by using Respiratory Syncytial Virus (RSV) Real Time RT-PCR Kit (Shanghai ZJ Bio – Tech Co., ltd) according to manufacturer’s instructions.
cDNA synthesis
This was accomplished by adding 0.1-1.0 ng / µl template RNA and RNase/DNase free water into the Maxime RT Pre-Mix tubes (Oligo dT15 or Random Primer) to a total volume of 20μl (iNtRON Biotechnology, Inc., Korea). cDNA synthesis that was carried off at 45°C for 60 minutes, and followed by Reverse transcriptase (RTase) inactivation at 95°C for 5 minutes.
Nested RT-PCR for RSV-A
The primer sequences of RSV-A (RSV-A External PCR RSVA-G513-F: AGTGTTCAACTTTGTACCCTGC, RSVA-F131-R:CTGCACTGCATGTTGATTGAT, and for Nested PCR RSVAG606-F: AACCACCACCAAGCCCACAA,RSVA-F22-: CAACTCCATTGTTATTTGCC) [14], Optimized PCR reaction for RSV cDNA amplification was performed as follows, 3µl of RNA extract was added to 5 µl PCR premix (Maxime PCR premix kit (i-Tag)) containing i-Tag TM DNA polymerase, dNTP mixture and reaction buffer. Two µl of primers and 15µl of distilled water were then added to the PCR premix. The PCR program consisted of 94°C for 5 min, followed by 40 cycles of PCR, consisting of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C, with a final extension step of 10 min at 72°C. The expected productsof583bpamplicon were analyzed by electrophoresis on a 1.5% agarose gel; the product was visualized by staining with 0.15% Ethidium bromide using UV gel documentation system INGeNius (Analytika Jena, Germany). For the nested PCR the cycling protocol was the same as for the external PCR, except for the annealing temperature, which was 53°C. The nested amplicons of 391 bp were visualized by agarose gel electrophoresis as mentioned above.
RT-PCR for RSV-B
Conventional RT-PCR Primers for RSV-B (BGFSeq.1: AGAGACCCAAAAACACYAGCCAA,BGRSeq.2: ACAGGGAACGAAGTTGAACACTTCA) [14], were used Optimized PCR reaction for RSV cDNA amplification was performed as follow, 3µl of RNA extract was added to 5 µl PCR premix [Maxime PCR premix kit (i-Tag)] containing i-Tag TM DNA polymerase, dNTP mixture and reaction buffer. Two µl of primers and 15µl of distilled water were then added to PCR premix. The PCR program consisted of the following protocol; 94°C for 15 min; 40 cycles of 94°C for 30 s, 63°C for 1 min, and 72°C for 1 min, and final extension step at 72°C for 10 min. The expected amplified amplicon of 772 bp were analyzed by electrophoresis and visualized as described above to PCR and nested PCR.
Detection of RSVs prevalence
Nasal swap out of the 92-sample examined by RT-PCR, 21 (22.8%) were positive for RSVs. Of them 20 were of type A and 2 were confection of type A and type B identified by nested PCR [Table 1 and 2, Figure 1 and 2].
Table 1. Frequency distribution of RSV type A and B positive infection. *P ≥ 0.05.
Age group |
Male |
Female |
RSV-A |
RSV-B |
0 – 1 year |
7 |
7 |
14 (7 male, 7 female) |
1 male |
≥ 1 – 2year |
4 |
3 |
7 (4 male, 3 female) |
1 female |
Total |
21 |
10 |
21 (11 male, 10 female) |
2 |
Table 2. Frequency distribution of RSV A and B according to gender. (%) was calculated from total number of positive patient. P ≥ 0.05.
Sex (total) |
RSV–A |
RSV-B |
Total |
Male (48) |
14 (15.2%) |
1 (2%) |
15 (16.3%) |
Female (44) |
7 (7.6%) |
1 (2.3%) |
8 (8.7%) |
Total (92) |
- (22.8)
|
2 (2.2) |
|
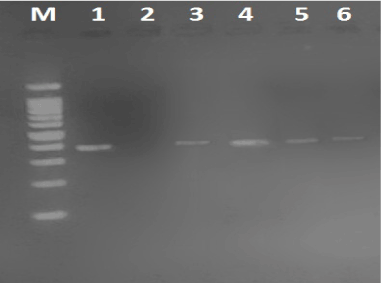
Figure 1. RSV-A result (391bp) on 2% agrose gel. lane1 shows positive control, lanes 2,4,5 and 6 show results in patients, lane 2 show negative control, M: 100bp DNA Marker.

Figure 2. RSV-B result (772 bp) on 2% agrose gel. lane1 shows negative control, lanes 2,3,4 ,5 and 6 show results in patients, lane 7 show positive control, M: 100bp DNA Marker.
RSV is a leading cause of bronchiolitis in infants and the elderly, and is estimated to cause 30 million lower respiratory tract infections, respiratory syncytial virus also represents a substantial burden of acute respiratory tract illness particularly in the early years of life leading to severe morbidity and hospitalization in children [15, 16].
In Sudan work on RSV is very scanty, nominal information is available about the incidence of RSV infection in infant; however, pneumonia was cause death in 2004, 2005 and 2006 in infants in Sudan [17]. In the present study, ninety two samples were collected from children with upper RTI mainly bronchiolitis and high rate of RSV infection was detected by RT-PCR; and 21(22.8%) were reported as positive cases. Similar results were recorded in Egypt in which, RSV was detected in 16.4% of the cases [18]. However, lower incidences were reported from Kenya where 166 out of 2143 (8%), and from India where 21 out of 200 (10%) were confirmed positive for RSV infection [19,20].
Our results also provide the first report on molecular detection of RSV genotypes in Sudan. In this study, only children suspected to be infected by RSV were included. The prevalence rate of RSV was 21 (22.8%) using real time PCR (RT-PCR), in age group < 2years, in which bronchiolitis was the most frequent clinical diagnoses, In comparison to RSV-B, a significant greater number of RSV-A positive cases was identified.
Our finding the prevalence’s of RSV- A in male (15.2 %) and in female (7.6 %), and RSV-B in male (2%) and in female (2%), in previous study revealed that, incidence of RSVA is more in children less than one year more. This may have expected by bronchiolitis is more severe in younger children [21].
The results obtained in this study show the need for wider surveillance at national level, in order to fully elucidate the true status of RSVs infection in Sudan. In this study, the use of RT-PCR in the detection of RSV among bronchiolitis children patients was established. This study was conducted to serve as a baseline for future plans aiming to study RSV in Sudan.
2021 Copyright OAT. All rights reserv
In summary, the incidence and existence of RSV in Sudan was documented through the detection of RSV indicating a high prevalence among bronchiolitis children patients in Sudan. Moreover, RSV genotypes detection using nested RT PCR was established. Generally, these findings are useful for future studies, since there is little information available about RSV infection in Sudan.
Our result showed that both RSV-A and RSV-B genotypes are co-circulatory in Sudan. The infection was found to be equally prevalent in both males and female children but significantly higher in younger infant (1 years) it recommended that wider use of vaccine should be practiced in children less than 2 years with bronchiolitis and morbidity in Sudan, and RSVA more commonly also carry wide surveys are needed to study RSVs infections and that genotypes distribution in Sudan.
The authors wish to thank Mohamed Alamin hamid pediatric hospital in Omdurman for permission to collect the samples.
Fatima Altayb Ebrahim the sample collection; Fatima Altayb Ebrahim and, Mohamed O. Mustafa conducted the RNA extraction and RT-PCR and nested RT-PCR assay; Khalid A Enan revised the PCRassay reaction and validation and contributed to the concept and design of the study; AbdAlrheem Mohamed Elhussein contributed to the revision.
Funding/Support: This study was supported by Central Laboratory, Ministry of Higher Education and Scientific Research, Sudan.
- Hall CB (2001) Respiratory syncytial virus and parainfluenza virus. N Engl J Med 344: 1917-1928. [Crossref]
- Hall CB, Douglas RG Jr (1981) Modes of transmission of respiratory syncytial virus. J Pediatr 99: 100-103. [Crossref]
- Cane PA1 (2001) Molecular epidemiology of respiratory syncytial virus. Rev Med Virol 11: 103-116. [Crossref]
- Gimferrer L, Campins M, Codina MG, Martín Mdel C, Fuentes F, et al. (2015) Molecular epidemiology and molecular characterization of respiratory syncytial viruses at a tertiary care university hospital in Catalonia (Spain) during the 2013–2014 season. J ClinVirol 66: 27–32.
- Malasao R, Okamoto M, Chaimongkol N, Imamura T, Tohma K, et al. (2015) Molecular characterization of human respiratory syncytial virus in the Philippines, 2012–2013. PLOS ONE 10: e0142192.
- Esposito S, Piralla A, Zampiero A, Bianchini S, Di Pietro G, et al. (2015) Characteristics and their clinical relevance of respiratory syncytial virus types and genotypes circulating in Northern Italy in five consecutive Winter seasons. PLOS ONE 10: e0129369.
- CDC (2008) Centres for Disease Control and Prevention RSV: infection and incidence.
- Langley GF, Anderson LJ (2011) Epidemiology and prevention of respiratory syncytial virus infections among infants and young children. Pediatr Infect Dis J 30: 510-517. [Crossref]
- Goto-Sugai K, Tsukagoshi H, Mizuta K, Matsuda S, Noda M, et al. (2010) Genotyping and phylogenetic analysis of the major genes in respiratory syncytial virus isolated from infants with bronchiolitis. Jpn J Infect Dis 63: 393–400.
- McLellan JS, Chen M, Chang JS, Yang Y, Kim A, et al. (2010) Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J Virol 84: 12236–12244.
- World Health Organization (WHO) (2008) Generic protocol to examine the incidence of lower respiratory infection due to respiratory syncytial virus in children less than five years of age.
- Henrickson KJ (2004) Advances in the laboratory diagnosis of viral respiratory disease. Pediatr Infect Dis J 23: S6-10. [Crossref]
- Empey KM, Peebles RS Jr, Kolls JK (2010) Pharmacologic advances in the treatment and prevention of respiratory syncytial virus. Clin Infect Dis 50: 1258-1267. [Crossref]
- Uzma BA, Muhammad MA, Hajra S, Syed SZZ, Birjees MK (2013) Molecular characterization of circulating respiratory syncytial virus (RSV) Genotypes in Gilgit Baltistan Province of Pakistan during 2011-2012 Winter Season. PLOS ONE 8: e74018.
- Venter M, Madhi SA, Tiemessen CT, Schoub BD (2010) Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J Gen Virol 82: 2117-2124.
- McLellan JS, Chen M, Kim A, Yang Y, Graham BS, et al. (2010) Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat Struct Mol Biol 17: 248-250. [Crossref]
- Anon (2008) Detection and Molecular Characterization of Respiratory Syncytial Virus (RSV) in Children with Respiratory Signs in Khartoum State, Sudan 2011-2012. Annual Reports of Ministry of Health, Sudan American Journal of Infectious Diseases and Microbiology 3: 6-13.
- Fattouh AM, Mansi YA, El-anany M, El-kholy AA, El-karaksy HM (2011) Acute lower respiratory tract infection due to respiratory syncytial virus in a group of Egyptian children under 5 years of age. Ital J Pediatr 37: 14.
- Okir EA, Ngama M, Bett A, Nokes DJ (2011) The incidence and clinical burden of respiratory syncytial virus disease identified through hospital outpatient presentations in Kenyan children. PLOS ONE 7: e52520.
- Maitreyi SR, Wayne MS, Prasad AK, Lalit D, Broor S (2003) Genetic variability among group A and B respiratory syncytial virus isolates from a large referral hospital in New Delhi, India. J Clin Microbiol 41: 2311.
- Borchers AT, Chang C, Gershwin ME, Gershwin LJ (2013) Respiratory syncytial virus--a comprehensive review. Clin Rev Allergy Immunol 45: 331-379. [Crossref]


