Abstract
Objective: Nodal disease (N+) for early-stage oral squamous cell carcinoma (OSCC) is the most significant prognostic factor for survival. There is a lack of effective predictor to justify prophylactic neck dissection. Quantitative tissue pathology (QTP) has shown its promise in providing an objective means for diagnosis and prognosis of many cancer types. We conducted a pilot study on the utilization of QTP to evaluate risk of nodal disease.
Study design: Retrospective case-control study
Subjects and methods: Histological sections from 15 primary tumors of clinically node-negative (N0) patients were stained with Feulgen-Thionin followed by acquisition of digital images and image processing to measure the mean and variance of nuclear phenotype and tissue architecture features from 45,253 nuclei of 45 tumor nests. Association between features and nodal disease outcome (N0 or N+) was investigated using nested analysis of variance adjusted by patient. Ability to discriminate between N0 and N+ was analyzed using multivariate logistic regression and receiver operating characteristics (ROC) curve analysis. P-value<0.05 (2-sided) was considered significant.
Results: The N+ group presented higher mean values of chromatin condensation levels and cell density compared to those of the N0 group. ROC curve showed a strong discriminative ability of chromatin condensation levels between the N+ and N0 groups with a sensitivity and specificity of 1.0 and 0.75, respectively.
Conclusion: This study reports the first-ever data on QTP as a risk assessment tool for nodal disease in early-stage OSCCs. Such computational imaging analysis potentially provides a new objective approach to predict regional nodal disease.
Key words
nodal disease, oral squamous cell carcinoma, elective neck dissection, tumour nests
Introduction
For patients with primary oral squamous cell carcinoma (OSCC) the most significant prognostic factor is the spreading of tumor to lymph nodes of the neck where the presence of tumor in any single node can reduce survival by 45-50% [1-3]. Thus, all neck nodes must be clinically assessed prior to treatment planning. For clinically node-negative (cN0) patients, most clinicians advocate elective neck dissection (END) to minimize the chance of tumour spread [4]. However, to avoid over-treating ~75% patients who would never develop nodal disease, some clinicians prefer to wait and dissect only when nodal disease actually occurs; and for tumors that eventually metastasize, patients may miss the window of early intervention leading to under-treatment or death. The decision on how to manage neck nodes remains controversial. In current practice, the decision of END is most often based on indicators from histological descriptions of the surgical samples. However, such pathology interpretation can be subjective and may often lead to over treatment [5,6]. In our population-based retrospective study, we found that tumor depth of invasion (DOI), a commonly used marker for the justification of END, is a poor indicator of nodal disease for early-stage cN0 OSCCs [7]. To avoid over treating those who will not develop nodal disease or under treating those who will benefit from early intervention, effective markers are urgently needed for the prediction of lymph node status in cN0 OSCC patients.
Quantitative tissue phenotyping (QTP) by computational image analysis has become a novel emerging means of obtaining objective information concerning diagnosis and prognosis of many cancer types [8-13]. During carcinogenesis, squamous cells go through progressive changes in appearance due to accumulation of genetic and epigenetic alterations, making them phenotypically different from normal cells. In our previous study, Guillaud et al. developed a scoring system (nuclear phenotype score, NPS) for premalignant lesions on cancer progression by recognizing nuclear phenotype that are distinctively different between normal, mild/moderate dysplasia, severe dysplasia/carcinoma in situ, or SCC [14]. We have also used this technique to assess surgical margins to predict the chance of recurrence [15]. In this case control study, we aim to use QTP to measure NPS of tumor nests of primary OSCC tumors of cN0 patients to investigate its potential to predict nodal status during follow-up.
Materials and methods
Patient and surgical specimen
From patients enrolled in the Canadian Optically-guided approach for Oral Lesions Surgical (COOLs) Trial [7,16,17], we identified 15 who had no previous history of OSCC, received surgical excision with curative intent, and were periodically followed up with thoroughly annotated information on demographics, clinic-pathological tumor characteristics, and clinical outcome on survival or nodal disease. The average age was 58.7±11.9 years and the male to female ratio was 1:1. Of the 15 patients, 8 remained N0 with a median follow-up time of 4.1 years, and 7 were found to be N+ after surgery or during follow-up. There was no difference in clinical or pathological attributes of the primary tumors between the two groups (Table 1).
Table 1. Patient demographics and clinic-pathological characteristics of primary tumors
Variables |
Total (N=15) |
N0 (N=8, 53%) |
N+ (N=7, 47%) |
P |
Age (years), mean ± SD |
58.7 ± 11.9 |
63.1 |
53.3 |
0.15 |
Age group |
|
|
|
0.26 |
<45 |
2 (13) |
|
2 (29) |
|
45-65 |
10 (67) |
6 (75) |
4 (57) |
|
>65 |
3 (20) |
2 (25) |
1 (14) |
|
Gender |
|
|
|
1.0 |
Male |
5 (33) |
3 (37) |
2 (29) |
|
Female |
10 (67) |
5 (63) |
5 (71) |
|
Smoking |
|
|
|
0.75 |
Never |
9 (60) |
4 (50) |
5 (71) |
|
Ever |
6 (40) |
4 (50) |
2 (29) |
|
Lesion site |
|
|
|
1.0 |
Buccal Mucosa |
1 (7) |
1 (12) |
|
|
Tongue / FOM |
14 (93) |
7 (88) |
7 (100) |
|
Tumor Grade |
|
|
|
0.13 |
I |
3 (20) |
|
2 (29) |
|
II |
11 (73) |
6 (75) |
4 (57) |
|
III |
1 (7) |
2 (25) |
1 (14) |
|
DOI (mm), mean ± SD |
6.1 ± 3.6 |
6.4 ± 4.5 |
6.7 ± 3.3 |
0.89 |
DOI of 4mm |
|
|
|
1.0 |
< 4mm |
2 (13) |
1 (13) |
1 (14) |
|
≥ 4mm |
13 (87) |
7 (87) |
6 (86) |
|
Survival status |
|
|
|
1.0 |
Alive |
11 (73) |
6 (75) |
5 (71) |
|
Dead due to any cause |
1 (7) |
|
1 (14) |
|
Dead due to OSCC |
3 (20) |
2 (25) |
1 (14) |
|
Years of follow-up, median (1st-3rd quartile) |
4.1 (2.8-5.0) |
41 (3.3-4.7) |
4.5 (2.3-5.0) |
0.96 |
N0 (node-negative); N+ (node-positive); FOM (floor of mouth); DOI (depth of invasion); OSCC (oral squamous cell carcinoma)
Two serial sections (4-µm thickness) from formalin-fixed paraffin-embedded (FFPE) primary tumors were obtained, one was stained with Hematoxylin and Eosin (HE) for visualizing cell type and tissue structure, and the other one was stained with Feulgen-Thionin (FT) (Figure 1A and 1B, respectively), using previously described protocol [18], for nuclear content.

Figure 1. Example of a tumor nest in QTP analysis. (A) A tumor nest stained with Hematoxylin & Eosin (H&E), (B) Feulgen Thionin (FT), and (C) an image (gray level) of the FT stain. (D). Subsequent segmentation generates Voronoi tessellation which constituted polygons in an arrangement of layers (e.g., a total of 11 layers for this nest). Polygons touching the nest outer boundary are depicted as Layer#1 (blue); Layer#2 (green) is made of polygons of immediate neighbors inner to Layer#1; and Layer#3 (red) is made of polygons of immediate inner neighbors touching Layer#2. Successive layers are incrementally calculated until there is no polygon left to be accounted for. (E) is the simplified cartoon of (D) to demonstrate layers and nest boundary (dashed line).
Image acquisition, segmentation and processing
Stained HE slides were imaged at 20X magnification with Pannoramic MIDI© and reviewed on Pannoramic Viewer© (3DHISTECH Ltd., Budapest, Hungary). Stained FT slides were imaged at 5 focal-planes every 500 µm using a modified version of our Cyto-Savant™ imaging system with bright-field microscopy (600 ± 5 nm illumination) and a charge-coupled digital camera which had a resolution of 1280 by 1024 (0.8 numerical aperture, effective pixel spacing in the sample plane was 0.27µm) [18].
Once digitized images were obtained, each pair of HE and FT images was put through an image analysis pipeline involving the following steps. First, an experienced pathologist (CFP) identified and demarcated tumor nests, the region of interests (ROIs), by drawing around the nest border on the HE images and matched the same ROIs on the FT images. Second, the best in-focus FT-stained image of each nucleus within each ROI was automatically selected and segmented using the DUnit Program© (Integrative Oncology, BC Cancer Agency, Vancouver, Canada) [14,19] (Figure 1C). Third, all segmented nuclei were manually evaluated and filtered to exclude: nuclei that were not squamous epithelial cells (granulocytes, lymphocytes, or fibroblasts); and ‘unacceptable’ nuclei that were overlapping with neighboring nuclei, incorrectly segmented, or out of focus. In the end, only nuclei of cancerous squamous cells were used for analysis.
QTP feature extraction
The DUnit program© automatically calculates 104 nuclear phenotype features and 16 tissue architectural features [20]. Nuclear phenotype features describe morphology, including the size, shape, and boundary variation of a nucleus based on the number of pixels (1 pixel = 0.116µm2) occupied by a nucleus; photometric based optical density for each pixel and chromatin texture evaluated using in fractal dimension calculations, Run_length, Markovian, and discrete texture features for each segmented nucleus. Tissue architecture was computed by constructing a Voronoi tessellation and Delaunay Triangulation, using the centers of gravity of the nuclei as seeds. Together, they measure the spatial distribution of cells and the distance between neighbouring cells thus portraying tissue organization. Mean and standard deviation of each feature were calculated within each ROI. The complete list of 120 features is given in Appendix Table 1.
Appendix
1. List of 120 measure quantitative tissue pathology (QTP) features
Phenotypes |
Category |
Sub-category |
QTP Feature Name* |
Nuclear |
Morphology |
|
Area (mean/stdv) |
Nuclear |
Morphology |
|
Mean_radius (mean/stdv) |
Nuclear |
Morphology |
|
Max_radisu (mean/stdv) |
Nuclear |
Morphology |
|
Var_radius (mean/stdv) |
Nuclear |
Morphology |
|
Sphericity (mean/stdv) |
Nuclear |
Morphology |
|
Eccentricity (mean/stdv) |
Nuclear |
Morphology |
|
Freq_low_fft (mean/stdv) |
Nuclear |
Morphology |
|
Freq_high_fft (mean/stdv) |
Nuclear |
Morphology |
|
Harmon01 (mean/stdv) |
Nuclear |
Morphology |
|
Harmon02 (mean/stdv) |
Nuclear |
Optical Density |
|
Dna_index (mean/stdv) |
Nuclear |
Optical Density |
|
Od_maximum (mean/stdv) |
Nuclear |
Optical Density |
|
Od_variance (mean/stdv) |
Nuclear |
Optical Density |
|
Od_skewness (mean/stdv) |
Nuclear |
Optical Density |
|
Od_kurtosis (mean/stdv) |
Nuclear |
Optical Density |
|
Var_intensity (mean/stdv) |
Nuclear |
Chromatin Texture |
Fractal Texture |
Fractal_dimen (mean/stdv) |
Nuclear |
Chromatin Texture |
Run Length |
Short_runs1 (mean/stdv) |
Nuclear |
Chromatin Texture |
Run Length |
Short_runs2 (mean/stdv) |
Nuclear |
Chromatin Texture |
Run Length |
Long_runs1 (mean/stdv) |
Nuclear |
Chromatin Texture |
Run Length |
Long_runs2 (mean/stdv) |
Nuclear |
Chromatin Texture |
Run Length |
Gray_level1 (mean/stdv) |
Nuclear |
Chromatin Texture |
Run Length |
Gray_level2 (mean/stdv) |
Nuclear |
Chromatin Texture |
Run Length |
Run_length1 (mean/stdv) |
Nuclear |
Chromatin Texture |
Run Length |
Run_length2 (mean/stdv) |
Nuclear |
Chromatin Texture |
Run Length |
Run_percent1 (mean/stdv) |
Nuclear |
Chromatin Texture |
Run Length |
Run_percent2 (mean/stdv) |
Nuclear |
Chromatin Texture |
Markovian Texture |
Entropy (mean/stdv) |
Nuclear |
Chromatin Texture |
Markovian Texture |
Energy (mean/stdv) |
Nuclear |
Chromatin Texture |
Markovian Texture |
Correlation (mean/stdv) |
Nuclear |
Chromatin Texture |
Markovian Texture |
Contrast (mean/stdv) |
Nuclear |
Chromatin Texture |
Markovian Texture |
Homogeneity (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Lowdnaarea (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Meddnaarea (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Hidnaarea (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Lowdnaamnt (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Meddnaamnt (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Hidnaamnt (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Lowdnacomp (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Meddnacomp (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Hidnacomp (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Mhdnacomp (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Low_av_dst (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Med_av_dst (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Hi_av_dst (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Mh_av_dst (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Lowvsmed_dna (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Lowvshigh_dna (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Lowvsmh_dna (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Low_den_obj (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
Med_den_obj (mean/stdv) |
Nuclear |
Chromatin Texture |
Discrete Texture |
High_den_obj (mean/stdv) |
Tissue Architecture |
Voronoi tessellation |
Density |
Density (mean/stdv) |
Tissue Architecture |
Voronoi tessellation |
VorArea |
VorArea (mean/stdv) |
Tissue Architecture |
Voronoi tessellation |
VorPeri |
VorPeri (mean/stdv) |
Tissue Architecture |
Voronoi tessellation |
VorRF |
VorRF (mean/stdv) |
Tissue Architecture |
Delaunay Triangulation |
DelNeiNb |
DelNeiNb (mean/stdv) |
Tissue Architecture |
Delaunay Triangulation |
NeaNei |
NeaNei (mean/stdv) |
Tissue Architecture |
Delaunay Triangulation |
DelNeaNei |
DelNeaNei (mean/stdv) |
Tissue Architecture |
Delaunay Triangulation |
Del3NeiDist |
Del3NeiDist (mean/stdv) |
*Each feature category or sub-category is described in its mean and variance.
Layering of tumor nests as ROI
In addition to investigating each tumor nest as a unit, we also performed layer-based analysis [21]. By generating Voronoi tessellation, which geometrically partitions each nest into regions based on the positions of the nuclei, we generated successive layers for each nest in the following manner. All nuclei whose corresponding Voronoi polygons touched the nest border were assigned to Layer#1 (the “outermost layer”); Layer#2 consisted nuclei which were not in Layer#1 and had a Voronoi neighbour in Layer#1; Layer#3 consisted nuclei which were not in Layer#1 or Layer#2 and had an neighbour in Layer#2. Higher number layers were defined similarly (Figure 1Dand 1F). We combined each layer with its subsequent layer for the purpose of obtaining more representative measurements considering: 1) the possibility that these tumor nests are tangentially sectioned and that a segmented nucleus from a 2-dimension image gives only partial phenotypic information of a whole nucleus; and 2) the less abundant well-segmented nuclei in the layers. For example, we treated Layer#1 and Layer#2 or Layer#2 and Layer#3 as a single ROIs (Layer#1-2 or Layer#2-3 respectively) by adding the nuclei from the 2 individual layers.
Statistical analysis
Each feature was compared between the N0 and N+ groups, with adjustment for patient-tumor nest effect by performing nested analysis of variance (nANOVA) tests. Distribution of each feature was illustrated by box-and-whisker plots for the two groups. For evaluating the ability of each feature in discriminating ROIs into the correct nodal status group, we first performed a forward stepwise linear discrimination process to select the feature(s) with the highest discriminative power. Due to the relatively large number of variables and small number of ROIs, we stopped the selection process at maximum of two features to avoid overfitting. Selected feature(s) were then tested for classification performance analysis by receiver operating characteristic (ROC) curve, with area under the curve (AUC) as a representation of the predictability for nodal status. For combinations of two selected features, a fitted probability of being N+ based on the model and features was computed. A threshold value with the highest sum of sensitivity and specificity was then calculated. All statistical analysis and plots were produced using R software (v3.2.3). As these were single variable and unpaired comparisons, all P values were uncorrected with P<0.05 considered to be statistically significant.
Results
A total of 45 tumor nests were identified with 23 (51%) from N0 and 22 (49%) from N+ groups (Table 2). These nests composed of 410 layers (N0, n=199; N+, n=211) and 45,253 segmented nuclei that were included for calculation of QTP features. There was no difference in the average number and pixel areas of the tumor nest per patient, the average number of layers per nest, or the average nuclei per nest between the two nodal status groups (Table 2).
Table 2. Tumor nests, layers, and cell nuclei and their lymph node status
Patient |
15 |
N0, 7 |
N+, 8 |
P |
Tumor nest (%) |
45 |
23 (51) |
22 (49) |
|
Tumor nest per patient |
3 |
2.9 |
3.1 |
|
Tumor nest area
(pixel area, µm2 ± SD) |
191,944.6 ± 256,088 |
238,211.3 ± 312,791.3 |
143,574.9 ± 173,536.8 |
0.22 |
Layers |
410 |
199 |
211 |
0.54 |
Layers per nest |
|
9.6 |
10.6 |
|
Cell nuclei |
45,253 |
22,996 |
22,257 |
0.97 |
Cell nuclei per nest |
|
999.8 |
1011.7 |
|
Abbreviation: N0, node-negative; N+, node-positive; SD, standard deviation
Association between QTP feature and lymph node status
Tumor nests as ROIs: Among the 104 nuclear phenotypic features, 6 discrete chromatin texture features were significantly different between the two groups; they all indicate a significant higher fraction of high or medium condensation level of chromatin regions in the N+ group (Table 3 and Figure 2).
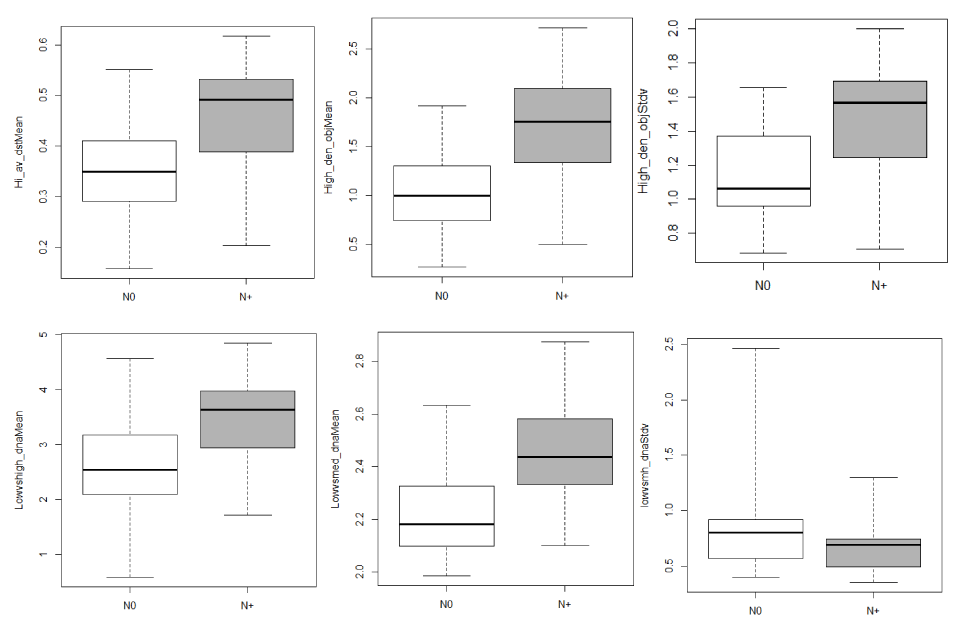
Figure 2. Box-and-whisker plots of nuclear phenotype features between N0 and N+ tumor nests. Top row, left to right: Hi_av_dstMean (P=0.04), High_den_objMean (P=0.01), High_den_objStdv (P=0.04). Bottom row, left to right: Lowvshgh_dnaMean (P=0.02), Lowvsmed_dnaMean
(P=0.003), Lowvsmh_dnaStdv (P=0.04). The horizontal line within the box indicates the median, the boundaries of the box (bottom and top) indicate the 1st and 3rd quartile, and the whiskers indicating the lowest and highest values.
Table 3. QTP features of significant difference between N0 and N+ tumor nests
| |
|
QTP Category |
QTP Sub-category |
Feature Name |
P |
Nuclear Phenotype (6) |
|
Chromatin Texture |
Discrete Texture |
Hi_av_dstMean |
0.04 |
|
Chromatin Texture |
Discrete Texture |
High_den_objMean |
0.01 |
|
Chromatin Texture |
Discrete Texture |
High_den_objStdv |
0.04 |
|
Chromatin Texture |
Discrete Texture |
Lowvshigh_dnaMean |
0.02 |
|
Chromatin Texture |
Discrete Texture |
Lowvsmed_dnaMean |
0.003 |
|
Chromatin Texture |
Discrete Texture |
Lowvsmh_dnaStdv |
0.04 |
Tissue Architecture (8) |
|
Delaunay Triangulation |
|
Del3NeiDistMean |
0.04 |
|
Delaunay Triangulation |
|
Del3NeiDistStdv |
0.007 |
|
Delaunay Triangulation |
|
DelNeaNeiMean |
0.03 |
|
Delaunay Triangulation |
|
DelNeaNeiStdv |
0.008 |
|
Voronoi tessellation |
|
VorAreaMean |
0.02 |
|
Voronoi tessellation |
|
VorAreaStdv |
0.007 |
|
Voronoi tessellation |
|
VorPeriStdv |
0.008 |
|
Voronoi tessellation & Delaunay Triangulation |
|
Density |
0.01 |
Eight tissue architecture features also showed significant differences between N0 and N+ groups (Table 3 and Figure 3). The N+ group had smaller nest area and circumference (VorAreaMean, VorPeriMean; P<0.05), and shorter distance between neighbours (DelNeaNeiMean, Del3NeiDistMean, P<0.05); collectively, these described smaller tumor nests and more densely packed cell nuclei (P=0.01).
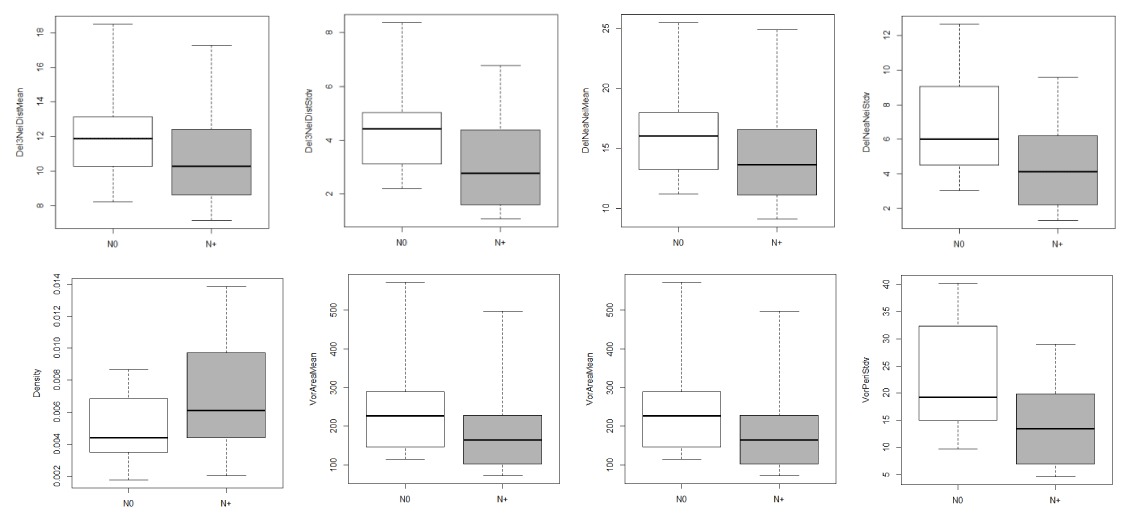
Figure 3. Box-and-whisker plots of tissue architecture features between N0 and N+ tumor nests
Top row, left to right: Del3NeiDistMean (P=0.04), Del3NeiDstStdv (P=0.007), DelNeaNeiMean (P=0.03), DelNeaNeiStdv (P=0.008). Bottom row, left to right: Density (P=0.01), VorAreaMean (P=0.02), VorAreaStdv (P=0.007), VorPeriStdv (P=0.008). The horizontal line within the box indicates the median, the boundaries of the box (bottom and top) indicate the 1st and 3rd quartile, and the whiskers indicating the lowest and highest values.
Combined tumor nest layers as ROIs: In Layer#1-2, the N+ group exhibited significantly higher fraction of medium and high density chromatin condensation states (Lowvsmed_dnaMean, P<0.001; Lowvshigh_dnaMean, P=0.03). There was also higher value in dispersion of chromatin of high condensation state in the N+ group (High_den_objMean, P=0.003) (Figure 4A).

Figu re 4. Box-and-whisker plots of nuclear phenotype features among combined layers.
a. Layer#1-2, top row, left to right: Lowvsmed_dnaMean (P<0.001), Lowvshigh_dnaMean (P=0.03), High_den_objMean (P=0.004); High_den_objStdv (P=0.02); bottom row, left to right: Run_length1Mean (P<0.01) and Run_length2Mean (P=0.03). b. Layer#2-3, left to right: Lowvsmed_dnaMean (P=0.01) and High_den_objMean (P=0.02). The horizontal line within the box indicates the median, the boundaries of the box (bottom and top) indicate the 1st and 3rd quartile, and the whiskers indicating the lowest and highest values.
In Layer #2-3, a similar observation was also seen where the N+ group had higher fraction of medium-density chromatin region (Figure 4B). We also observed higher average values in run_length features which describe the size of chromatin clumps where the larger the run_length values, the bigger the clumps and vice versa.
Discriminative ability on nodal status
The observed differences in the features described above provided initial data supporting QTP analysis for differentiating N+ from N0. Thus, we next investigated whether these 120 features can be used to predict N+ status. Linear discrimination analysis with forward stepwise selection procedure was performed to determine feature(s) that achieved the best discriminatione between the two groups among a) tumor nests and b) combined layers.
For both types of ROIs, feature selection from the 120 features consistently isolated Lowvsmed_dnaMean as the most independent discriminative feature. For tumor nests, combination of Lowvsmed_dnaMean and VorPeriStdv gave an AUC of 85%. The threshold value of fitted probability based on the model on selected features was 0.41 with sensitivity of 82% and specificity of 78%. When the combination of Lowvsmed_dnaMean and Mh_av_dstStdv were used, combined Layer #1-2 showed AUC of 94% (Figure 5) with sensitivity of 100% and specificity of 75%.
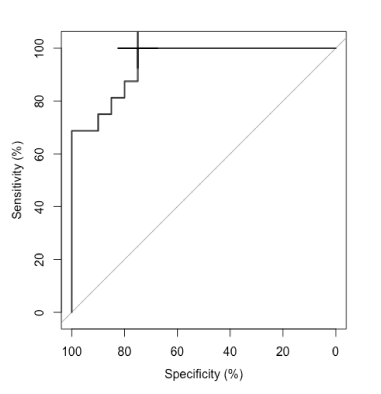
Figure 5. Receiver operating characteristic (ROC) curve for discriminating N+ from N0
among Layer#1-2. Combination of lowvsmed_dnaMean and Mh_av_dstStdv achieved area under the curve of 94% with threshold of fitted probability at 0.28 (black cross) achieved sensitivity of 100% and specificity of 75%.
Discussion
Assessment and judgement of risk for nodal metastasis has always been a challenge for clinicians when facing cN0 OSCC patients. With its robust and objective nature, we report the first-ever use of QTP for quantitative analysis of tumour nests for predicting nodal disease. Comparison of QTP features between the two groups highlighted a reoccurring theme where the N+ nests demonstrated a higher ratio of medium to low condensation regions of chromatin and high cell density.
Nuclear chromatin alteration has been extensively studied as one of the regulators for genomic activity and functions which lead to aberrant chromatin remodeling that is seen in many human diseases and cancers [12,22-24]. During normal cell cycle, the chromatin de-condenses and becomes loosely packed euchromatin, exposing DNA and allowing gene activity and transcription. When activity is not needed, chromatin becomes tightly packed condensed heterochromatin. Taken together, changes in condensation states of chromatin occur throughout cell division. Therefore, it could be hypothesized that the observed higher fraction of medium-condensed chromatin reflects higher proliferation activity in N+ tumor cells.
The intention for studying and comparing nest layers was motivated by the increasing evidence on the role of tumor microenvironment in initiation, growth, and spread of tumor. Our study found that medium-to-low condensation ratio was significantly higher in nuclei of the outer layers of N+ tumor nests than that of the N0 group. Tumor nests are bordered by a well-defined boundary; and immediately inside this boundary are most likely where proliferative tumor cells reside. Indeed, the progression to cancer and metastasis require not only genomic and morphological changes but also cascades of events and responses with local microenvironment leading to loss of polarity, disruption in epithelial compartment, invasion, and angiogenesis [25-28]. This particular finding could indicate an interaction of these outer layer cells with tumor microenvironment which may play important roles in nodal metastasis. The current QTP system has the capacity to analyze in situ-stained specimens. From this point and moving forward, we plan to integrate tumor QTP features with profiles of surrounding stromal cells, including inflammatory cell types, especially for areas at the invasive front. This may help us explain the higher state of chromatin condensation seen in N+ group as well as elucidate the possible anti-cancer or pro-cancer interaction between the N+ and tumor microenvironment.
Regarding the tissue architecture of these tumor nests, we observed a strong correlation between cell density and nodal status, as characterized by smaller Voronoi polygon area and shorter distance between neighbour nuclei. From this, we attempted to infer NC (nucleus-cytoplasm) ratio, a measure commonly used in the conventional pathology, by associating cytoplasmic area with Voronoi polygon area. In normal cells, the nuclei size decrease as the cell matures; thus the NC ratio decreases as well. This suggests that an increased NC ratio in matured cells indicate atypical growth pattern which is the general observation in premalignant or malignant cells [29]. In our pilot cohort, the average nuclear area did not differ between N+ and N0 groups; however, the average polygon area measured was significantly smaller in N+ group. If we take the N0 group as the reference, i.e., set the reference NC ratio to 1, to indicate non-metastatic potential, a smaller cytoplasm area with the same nuclear area would give a larger NC ratio. Analogously, we can infer that NC ratio with nodal status. Indeed, we observed N+ group had an increase in NC ratio in the outermost layers, but the difference was not significant (P=0.11).
In addition, fractal dimension (FD), which measures the extent of irregularity and complexity in nuclear structure has been associated with malignant, less differentiated tumors, and poor prognosis in many cancer types [10, 30-34]. Although there was no difference in FD between N0 and N+ groups, we observed a higher mean of FD in nuclei of poorly differentiated tumors. Thus, this feature may potentially be used to compensate the subjectivity in determining tumor differentiation which could be associated with nodal disease outcome.
The most important finding of our study is however, the value of QTP analysis as a predictive and risk assessment aid in pathology. Two features, both describing chromatin texture, demonstrated outstanding ability discriminating N+ from N0 nest layers with AUC of 94%. This is much better compared to using the combination of differentiation and DOI,[7] which has shown 63% of AUC with 67% specificity and 54% sensitivity.
This pilot study is li2021 Copyright OAT. All rights reserv included. Although there was no difference in demographics and clinic-pathological characteristics, the observed association and the predictive ability of QTP features requires further validation in a larger sample size cohort with prospective recruitment, such as the COOLS study [16]. Nevertheless, despite the small number of patients, the number of tumor nuclei and layers of tumor nests are enough for meaningful statistical results.
In conclusion, the results of our study encourage the potential value of bringing computational imaging analysis as an adjunct tool for pathologists in assessing patients who may be at high risk of developing nodal disease. Additional studies are warranted with increased number of patients to corroborate the findings provided in this work.
Acknowledgments
The work was supported by the BC Cancer Foundation and UBC Oral Cancer Research Fund. The authors would also like to thank the following support from the Department of Integrative Oncology, British Columbia Agency/Research Center, and Vancouver, British Columbia, Canada: Monica Ye for statistical support, Jagoda Korbelik, Cindy Cui, and Anita Carraro for their technical support.
References
- Mamelle G, Pampurik J, Luboinski B, Lancar R, Lusinchi A, et al. (1994) Lymph node prognostic factors in head and neck squamous cell carcinomas. Am J Surg 168: 494-498. [Crossref]
- Layland MK, Sessions DG, Lenox J (2005) The influence of lymph node metastasis in the treatment of squamous cell carcinoma of the oral cavity, oropharynx, larynx, and hypopharynx: N0 versus N+. Laryngoscope 115: 629-639. [Crossref]
- Yuasa-Nakagawa K, Shibuya H, Yoshimura R, Miura M, Watanabe H, et al. (2013) Cervical lymph node metastasis from early-stage squamous cell carcinoma of the oral tongue. Acta Otolaryngol 133: 544-551. [Crossref]
- Liu KY, Durham JS, Wu J, Anderson DW, Prisman E, et al. (2016) Nodal Disease Burden for Early-Stage Oral Cancer. JAMA Otolaryngol Head Neck Surg. [Crossref]
- Bryne M, Nielsen K, Koppang HS. Dabelsteen E (1991) Reproducibility of 2 malignancy grading systems with reportedly prognostic value for oral-cancer patients. J Oral Pathol Med. 20: 369-372. [Crossref]
- Russolo M, Giacomarra V, Papanikolla L. Tirelli G (2002) Prognostic indicators of occult metastases in oral cancer. The Laryngoscope. 112: 449-452. [Crossref]
- Liu KYP, Durham JS, Wu J, et al (2016) To dissect or not to dissect the necks of oral cancer patients with clinically negative nodal disease? In JAMA Otolaryngology–Head & Neck Surgery.
- Mulder JW, Offerhaus GJA, Defeyter EP, Floyd JJ, Kern SE, et al. (1992) The relationship of quantitative nuclear morphology to molecular genetic alterations in the adenoma-carcinoma sequence of the large-bowel. Am J Pathol. 141: 797-804. [Crossref]
- Dong F, Irshad H, Oh EY, Lerwill MF, Brachtel EF, et al. (2014) Computational pathology to discriminate benign from malignant intraductal proliferations of the breast. PLoS One 9: e114885. [Crossref]
- Tambasco M, Eliasziw M, Magliocco AM (2010) Morphologic complexity of epithelial architecture for predicting invasive breast cancer survival. J Transl Med 8: 140. [Crossref]
- Wang LW, Qu AP, Yuan JP, Chen C, Sun SR, et al. (2013) Computer-based image studies on tumor nests mathematical features of breast cancer and their clinical prognostic value. PLoS One. 8: e82314. [Crossref]
- Vanvelthoven R, Petein M, Oosterlinck WJ, Roels H, Pasteels JL, et al. (1995) The use of digital image-analysis of chromatin texture in feulgen-stained nuclei to predict recurrence of low-grade superficial transitional-cell carcinoma of the bladder. Cancer. 75: 560-568. [Crossref]
- Rosin MP, Poh CF, Guillard M, Williams PM, Zhang L, et al. (2007) Visualization and other emerging technologies as change makers for oral cancer prevention. Ann N Y Acad Sci 1098: 167-183. [Crossref]
- Guillaud M1, Zhang L, Poh C, Rosin MP, MacAulay C (2008) Potential use of quantitative tissue phenotype to predict malignant risk for oral premalignant lesions. Cancer Res 68: 3099-3107. [Crossref]
- Ye M, Liu KYP, Poh CF, MacAulay CE. Guillaud M (2014) In Automated quantitative nuclear phenotype (anps) associates degree of dysplasia at surgical margins and fluorescent margins, Terry Fox Research Institute Annual Scientific Meeting, Vancouver, British Columbia, Canada, 2014; Vancouver, British Columbia, Canada.
- Poh CF, Durham JS, Brasher PM, Anderson DW, Berean KW, et al. (2011) Canadian optically-guided approach for oral lesions surgical (cools) trial: Study protocol for a randomized controlled trial. BMC cancer. 11: 462. [Crossref]
- Poh CF, Anderson DW, Durham JS, Chen J, Berean KW, et al. (2016) Fluorescence Visualization-Guided Surgery for Early-Stage Oral Cancer. JAMA Otolaryngol Head Neck Surg 142: 209-216. [Crossref]
- Garner D, Ferguson G. Palcic B (1994) The cyto-savant system. In Automated cervical cancer screening, Grohs HK, Husain, O, Eds. Igaku-Shoin. New York 305-317.
- Kamalov R, Guillaud M, Haskins D, Harrison A, Kemp R, et al. (2005) A Java application for tissue section image analysis. Comput Methods Programs Biomed 77: 99-113. [Crossref]
- Doudkine A, Macaulay C, Poulin N, Palcic B (1995) Nuclear texture measurements in image cytometry. Pathologica 87: 286-299. [Crossref]
- Guillaud M, Buys TP, Carraro A, Korbelik J, Follen M, et al. (2014) Evaluation of HPV infection and smoking status impacts on cell proliferation in epithelial layers of cervical neoplasia. PLoS One 9: e107088. [Crossref]
- Young IT, Verbeek PW, Mayall BH (1986) Characterization of chromatin distribution in cell nuclei. Cytometry 7: 467-474. [Crossref]
- Gao F, Jütting U, Rodenacker K, Gais P, Lin PZ (1997) Relevance of chromatin features in the progression of esophageal epithelial severe dysplasia. Anal Cell Pathol 13: 17-28. [Crossref]
- Cherkezyan L, Stypula-Cyrus Y, Subramanian H, et al. (2014) Nanoscale changes in chromatin organization represent the initial steps of tumorigenesis: A transmission electron microscopy study. BMC Cancer. 14: 189. [Crossref]
- Bissell MJ, Hines WC (2011) Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med 17: 320-329. [Crossref]
- Quail DF, Joyce JA (2013) Microenvironmental regulation of tumor progression and metastasis. Nat Med 19: 1423-1437. [Crossref]
- Deryugina EI, Quigley JP (2006) Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev 25: 9-34. [Crossref]
- Albinger-Hegyi A, Stoeckli SJ, Schmid S, Storz M, Iotzova G, et al. (2010) Lysyl oxidase expression is an independent marker of prognosis and a predictor of lymph node metastasis in oral and oropharyngeal squamous cell carcinoma (oscc). Int J Cancer. 126: 2653-2662. [Crossref]
- Mehrotra R, Gupta A, Singh M, Ibrahim R (2006) Application of cytology and molecular biology in diagnosing premalignant or malignant oral lesions. Mol Cancer 5: 11. [Crossref]
- Bose P, Brockton NT, Guggisberg K, et al. (2015) Fractal analysis of nuclear histology integrates tumor and stromal features into a single prognostic factor of the oral cancer microenvironment. BMC Cancer. 15.
- Mincione G, Di Nicola M, Di Marcantonio MC, Muraro R, Piattelli A, et al. (2015) Nuclear fractal dimension in oral squamous cell carcinoma: a novel method for the evaluation of grading, staging, and survival. J Oral Pathol Med 44: 680-684. [Crossref]
- Delides A, Panayiotides I, Alegakis A, Kyroudi A, Banis C, et al. (2005) Fractal dimension as a prognostic factor for laryngeal carcinoma. Anticancer Res 25: 2141-2144. [Crossref]
- Oczeretko E, Juczewska M. Kasacka I (2001) Fractal geometric analysis of lung cancer angiogenic patterns. Folia Histochem Cytobiol. 39 Suppl 2: 75-76. [Crossref]
- Dey P, Rajesh L (2004) Fractal dimension in endometrial carcinoma. Anal Quant Cytol Histol. 26: 113-116. [Crossref]





