Advances in Genetics led to the development of the Sciences of Omics and System Biology. They provide the tools for a better understanding of human diseases and for the development of new drugs and ultimately the possibility of Personalized Medicine. It is now possible to determine the entire DNA sequence of a genome as well as the entire protein sequence of a proteome in any organism because of the coming of Throughput technologies and Bioinformatics.
Thus, an understanding of DNA and Proteins is very important in maintaining the human health. The fact that gene makes protein which catalyzes biochemical reaction and controls the phenotype of organism has led to coming of therapeutic managements of medical conditions like providing insulin to diabetes patients who lack the ability to produce insulin due to a defective gene required for metabolism of sugar or controlling the intake of certain amino acids in patients with mental disorders like phenylketonuria and taking care of several other human diseases.
Besides catalyzing a metabolic reaction, proteins are important. Since they are also used as drugs to treat many diseases. In addition, several drugs interact with protein determining the effectiveness of the drugs and their side effects. Thus, understanding of DNA can identify the genes and protein responsible for diseases and for drug interaction
In view of these facts, the development of the science of Omics such as Genomics, Proteomics, Epi-genomics and Metabolomics has become very crucial for understanding the cause and the treatment of diseases and for the maintenance of human health. Understanding of the fact that a gene makes protein which catalyzes biochemical reaction and controls the phenotype of organism has led to the coming of therapy of medical conditions like providing insulin to diabetes patients or controlling the intake of certain amino acids in patients with mental disorders like phenylketonuria and taking care of several other human diseases.
Methods are now developed to decipher the entire sequence of the genome of a person within few hours in a cost-effective way: Which can be utilized to find the nature of a defective gene that is responsible for causing the disease; this paves the way for the development of Genomic medicine. The structure and function of Proteins became amenable to analysis after the establishment of relation between a gene and a protein by Beadle and Tatum in 1941 and the development of method for sequencing of amino acids in a protein by Edman in 1950.
Some of these aspects of Proteomics including the development of different genetic, biochemical and throughput technologies and future medical possibilities are very well established by now.
Hans Winkler in 1920 coined the term genome to include the haploid chromosome complement of an organism, much later in mid nineteen seventies MCKussic and Ruddle introduced the term genomics derived from Genome [1]. Later proteome was introduced to include the entire protein profile of an organism and the term proteomics was derived from proteome. However, Nobelist Professor Joshua Lederberg (2000) has suggested that the word Omics originated from Sanskrit word Om which encompasses the complete description of a system or universe.
The science of omics includes followings:
Genomics (complete nucleotide sequence i.e. genetic makeup of an organism)
Proteomics (complete proteins of a cell in any organism)
Epi-genomics (modification of nucleotides in an organism)
Metabolomics (Changes in the gene activity in response to metabolites)
Later Lederberg coined the term Microbiome to describe the microorganisms that exist in our guts and in that of other animals. Metagenomics is the genomics of microbiome. The significance of microbiome is revealed by the recent findings that certain changes in microbiome can lead to loss of motor function underlying the major symptom of Parkinson's Disease.
In this list, a few other kinds of omics may be added such as transcriptomics, interactomics, and metagenomics. Transcriptomics includes the description of transcripts; interactomics includes the description of interactions among proteins and metagenomics includes the genomics of micro-organisms resides within us or within other organisms.
Major Advances in Biology that led to coming of the Sciences of Omics includes followings. These advances integrate different branches of Biology and interconnect Biology with Chemistry and other Branches of Science. Some of these advances includes:
1. Mendel’s law of inheritance: This established the particulate nature of the gene.
2. One- Gene - One -enzyme Concept of Beadle & Tatum: This concept inter-relates Genes (DNA) to Protein and Biology to Chemistry via genetic control of Biochemical reactions.
3. Watson & Crick structure of DNA: This provided the molecular Basis of life and mechanism for: DNA replication, transfer for genetic information (genetic code, transcription & translation) and changes in Genetic information (Mutation).
4. Development of Different Technologies including Recombinant DNA technology/Cloning, Protein & DNA Sequencing and PCR and Throughput Technologies; gene-editing/ CRISPR and Bioinformatics.
Genomics i.e., complete nucleotides of an organism was made possible by development of technology for cloning genes, ability to amplify it by polymerase chain reaction (PCR), ability to sequence DNA base pairs by Sanger’s methods and its further modification and adaptation to throughput methods and finally assembly and annotation of DNA sequence by application of methods of Bioinformatics.
The entire DNA of an organism can be thus sequenced and this is called Whole Genome Sequence (WGS) or alternatively the DNA sequence of an organism can be derived after sequencing only a part of it which includes only the protein coding sequences: This method is called Exosome Genome Sequencing (EGS). EGS is much faster and less expensive even though not quite accurate.
The size of the genome of a large number of organisms are now established; some of them are listed in (Table 1). The cost of Genome sequencing has gone down drastically from 3 billion dollars for the first human genome sequencing in the beginning of this century to about 3 thousand dollars at present in the year 2016. The cost of DNA sequencing is expected to even further down to about one thousand dollars. Such drop-in sequencing cost will make it possible to allow routine use as diagnostic tool as other diagnostic technologies for example MRI. Such routine use of DNA sequencing will advance personal medicine. Now using the current technology, it is possible to sequence the DNA from a single cell: This will be very helpful in cancer genomics.
Table 1. Genomics of different organisms
Organism |
Year |
Size |
Comment |
fx174 virus |
1977 |
3 Kb |
First bacterial virus, not a free-living organism, sequenced |
Hemophilus influenze |
1995 |
1.8 Mb |
First bacterium or a free-living organism sequenced |
Sachcaromyces cerevisiae |
1996 |
12 Mb |
Yeast, the first eukaryote sequenced |
Ceonoribditis elegans |
1998 |
100 Mb |
Soil worm |
Drosophila melanogaster |
2000 |
180 Mb |
Fruit fly |
Human |
2000 |
3000 Mb |
The first draft of human genome sequence announced |
Chimpanze |
2005 |
3000 Mb |
The closet living relative of human |
*Neanderthal |
2006 |
3000 Mb |
The extinct cousin of human, over a million base pairs sequenced in 2006 but now fully sequenced. |
(After Mishra 2010 with permission of John Wiley & sons)
*Neanderthals, the closest primate is considered a human cousin. The human HAR genes are the stretch of DNA human genome which have undergone rapid mutations that’s the reason they are called Human Accelerated Regions.
Use of current DNA sequencing has led to the establishment of the genome sequences of over thousand species of organisms. Such knowledge of genome sequence has been useful in many ways. This led to understanding that the minimum number of genes to sustain life by a bacterium is only 260. It also led to surprising understanding humans carry only 23000 genes as against to previous guess that human may have up to one hundred thousand genes. Such establishment of meagre 23thousand genes has emphasized the role of RNA Processing in creation of multiple mRNA per gene. Comparisons of human genome sequences from different individuals as led to establishment of copy number variation (CNV) with over 10 million single nucleotide polymorphisms (SNP).
Knowledge of human DNA sequences and its comparison with other organisms particularly primates including Neanderthals makes it obvious that at least the presence of several genes makes us human and set human apart from others including Neanderthals. In human, there are at least 6 genes or DNA sequences including: Har1 controlling the development of cerebral cortex, FoxP2 endowing the ability of speech to human, Amy1 responsible for digestion of starch and sequence ASPM which controls the human brain size which has triple over the period of human evolution. Also, DNA sequence LCT which provides ability to digest milk sugar and use animal milk as additional source of food and DNA sequence HAR2 which permits dexterity of wrist &thumb and use of complex tools.
Knowledge of DNA sequence also provides a basis for the understanding of metabolic pathways and inherited disease: And above all a basis for the coming of Synthetic Genetics/Biology. Synthetic genetics has been possible because of our ability to edit DNA sequences. There are several ways to edit or manipulate DNA sequences, among them the most rapid one is the use of CRISP method that allows specific changes to be introduced at a specific site in the DNA
Proteomics is all about the proteins, their structure, function and interactions through which DNA controls the features of cells in any organism. Only a small number of genes or DNA sequences are transcribed and translated to yield protein. Some other DNA sequences are transcribed and /or then translated to yield products which facilitate the transcription of a gene and in translation of the transcript into proteins.
Protein controls the structure and function of a cell by facilitating all biochemical reactions. In addition, the significance of proteins in design of drugs becomes more important as a number of proteins act as drugs or interact with pharmaceutical drugs in human body. The different kinds of proteins are listed in (Table 2).
Table 2. Function of different proteins
S.NO |
Function |
Protein |
1 |
Catalyst |
Enzymes (over 90 % of proteins) Catalyze biochemical reactions in the cell |
2 |
Transport |
Hemoglobin (carrier of Oxygen) Albumin (carrier of hormones) |
3 |
Structure |
Cartilage / Bone proteins |
4 |
Cellular skeleton |
Actin, Fibrinoactin |
5 |
Hormone |
Insulin, Growth Hormone |
6 |
Antibody |
Immunoglobulins |
7 |
Antigens & allergens |
Bacterial & Viral proteins |
8 |
Mobility / muscle movement |
Myosin |
9 |
Receptors |
Receptor for Cholesterol |
10 |
Cell communication / Signaling |
Transduction proteins, Junction proteins |
(After Mishra 2010 with permission of John Wiley & Sons)
Proteomics first started with functional analysis of proteins based on one-gene-one-enzyme concept that a mutant or diseased organism must possess a defective or missing protein corresponding to a defective gene or DNA sequence
Proteomics first started with functional analysis of proteins based on one-gene-one-enzyme concept that a mutant or diseased organism must possess a defective or missing protein corresponding to a defective gene or DNA sequence [2] as shown below in case of hemoglobin of a person suffering from sickle cell anemia.
1 2 3 4 5 6 7 8
Hemoglobin A Val-His-leu-Thr-Pr0-Glu-Glu-Lys-
Hemoglobin S Val-His-Leu-Thr-Pr0-Val-Glu-lys-
And then study of proteomics was followed with the structural analysis of protein by methods of Edman’s degradation in early 1950s. Thus, former provided the tools to analyze the function of a protein whereas Edman’s method provided the structure of protein by determining the sequence of amino acids in a peptide.
Later proteomics included the chemical analysis of the structure of individual proteins by Throughput methods such as 2 D gel and mass spectroscopy as described later in this section.
Protein is a stretch of amino acids in which adjacent amino acids are joined by peptide bonds. Each protein has an N-terminal end and C-terminal end corresponding to the 5’end and 3’end of the nucleotide in the nucleotide sequence of the gene from which this protein is translated. Most proteins have a stretch of 10-15 amino acids on the N’-Terminal called as signal peptide which guides each protein to final destination or location: For example some proteins remain in the cell or in different organelles whereas other proteins are expelled to the exterior of a cell (Figure 1).

Figure 1. Protein Structure (Kindly Provided by Derryl Leza of NGHRI)
A protein can assume four levels of organization. These are known as primary, secondary, tertiary and quaternary structures or level of organization of a protein as depicted in the (Figure 2) below, kindly provided by Derryl Leza of NGHRI. The structure and function of protein was elucidated by the work of several biochemists like Stanford Moore, William Stein, Bruce Merrifield and Gunter Blobel [3,4] of the Rockefeller University and by Christen Anfinsen of the National Institute of Health for which they received Nobel prizes Much earlier the relation between gene and protein in controlling the metabolic roles was established by the Nobel Prize winners George Beadle and Edward Tatum. The process by which the structure of a protein is deformed is called denaturation whereas the reverse process by which proteins assume native structure is called renaturation. Now methods are known by which a protein can be denatured or renatured: For example, a boiled egg can be renatured to yield to the status of an un- boiled egg.
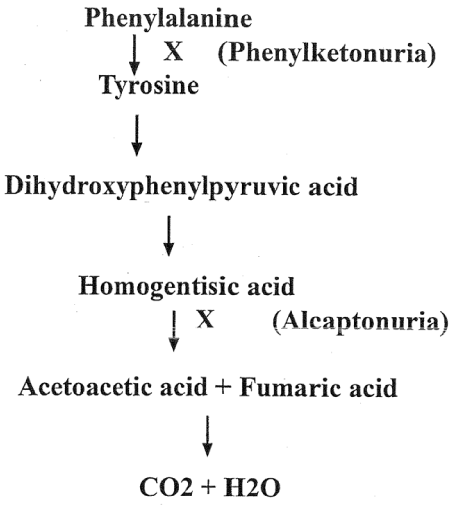
Figure 2. Metabolism of Phenylalanine in human
Understanding of the protein and its correlation with a gene. The one gene one enzyme concept provided the biochemical basis for understanding of the Gorrod’s [5] view of the inborn error of metabolism. It also served the tool for examining the metabolic pathway and their genetic control by comparing the defective protein (s) from the normal and patients. This method of Beadle and Tatum has been used every time for such analysis to this date.
Genetic Basis of Human Disease as depicted by metabolism of phenylalanine in human causing mental disorder and several other diseases as shown in the parenthesis of the metabolic flow chart below (Figure 2).
Understanding of the structure of gene and that of the protein encoded by this gene led to the establishment of Co-linearity of gene and protein. Co-linearity shows that if a nucleotide is changed in the codon of a gene them there must be a corresponding change in the amino acid in the protein tryptophane synthetase of Neurospora crassa [6] (Figure 2).
Chemical Analysis of Amino Acid Sequence in Protein by Edman Degradation by removal of one amino acid from N-terminus giving the entire sequence of the protein.
A Swedish Scientist Pehr Edman [7] while visiting the Rockefeller University developed a method in early 1950s to sequence amino acids from the N-terminal end one by one. In this method the N-terminal amino acid was labeled with a chromogenic chemical, Phenyl -isothiocyanate and then the peptide was subjected to mild hydrolysis which released the marked N-terminal amino acid which was subsequently identified after chromatographic separation and the cycle of labeling the N-terminal amino acid in the already shortened peptide and its mild hydrolysis was repeated until all the amino acids in sequence in the peptide were identified as depicted in the following diagram. This process is called Edman’s degradation. This process was later fully automated by Edman in Sydney, Australia. The method of Edman degradation was used by sanger in England in mid 1960s to determine the entire amino acid sequence of the two peptide chains in insulin. Soon after Edmans degradation was used by Moore and Stein in the USA to sequence the first large protein, Ribonuclease.
N’- amino acid1- amino acid 2- amino acid3- amino acid 4-amino acid5- C’
+ phenyl isothiocynate
Mild hydrolysis
Phenyl-isothiocynate-N’-amino acid1 + N’amino acid 2-aminoacid 3 – aminoacid 4 aminoacid5-C’
Proteomics flourished with the development of Throughput technology such as:
A. 2D Gel electrophoresis
B. Mass Spectrometry.
2D Gel electrophoresis: A Two-dimensional gel electrophoresis method that could separate hundreds of proteins on one gel was developed by O’Farrel [8] and by Klose [9] Simultaneously and independently. O’Farrel showed that over 1100 proteins from Escherichia coli cell extract could be separated by electrophoresis and visualized on one gel which are run in succession, the proteins are separated first based on their charge on an ampholine containing gel and then by mass on a gel containing sodium deodyl sulfate. Such two-dimensional gels (2D gel) are now routinely used to separate a large number of proteins from any cell / tissue or organism at once. The 2D gels are also used to compare the protein profiles of normal cells with that of the patient with a particular disease.
From such comparisons of protein profile on 2D gel the proteins responsible for diseased condition can be readily identified and then further characterize to reveal the molecular change in the proteins in a patient. A comparison of proteins from the cerebrospinal fluid of patients suffering from Alzheimer disease is presented in the figure below. The altered proteins in the Alzheimer patient are identified by change in their mobility and intensity (Figure 3 and 4).
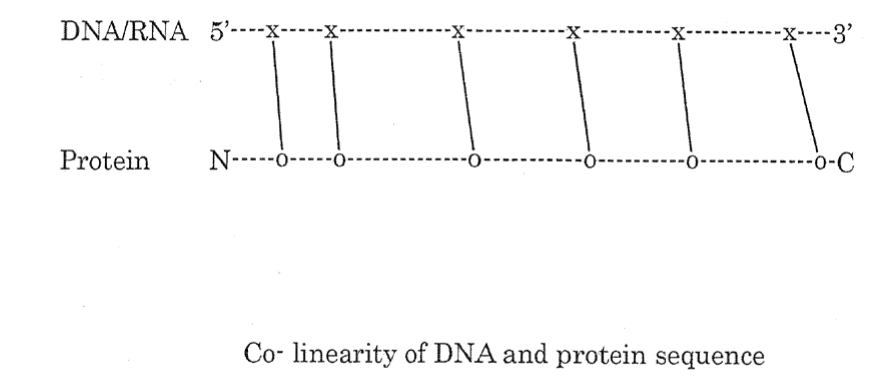
Figure 3. Co-linearity of DNA and Protein sequence
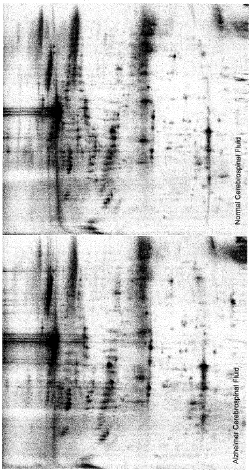
Figure 4. 2D gel separation of proteins of spinal fluid from normal and Alzheimer patient (Courtesy of Professor Kevin Lee, University of Delaware)
Mass Spectrometry: Mass Spectrometer (MS) a very sophisticated instrument was developed over several decades starting from the day of JJ. Thomson, the father of the science of Particle Physics [10]. The development of Mass Spectrometer involved a large number of physicists, engineers and other technical personel; several of them were awarded Nobel Prizes for their contribution. Fenn [11,12] developed the Eletro spray Ionization (ESI) whereas Tanaka [13] developed the MALDI method of ionization for which they shared Nobel Prize, these methods became suitable for the analysis of biological materials including proteins by mass spectrometry. Proteins of interest are further characterized by Mass Spectrometry to identify a protein by its amino acid sequence and to determine any molecular change between the protein from the normal individual and the patients or mutants. Routinely a protein of interest is isolated from the gel sliced out from the 2D-gel and then digested by trypsin or other proteolytic enzymes.
These fragments are further separated by electrophoresis or chromatography and then loaded onto a platform of the Mass Spectrometer.
These are ionized (by EPI or preferably by MALDI) and fragmented and then detected on the detecting device of the mass spectrometer. The mass of the protein /peptide fragments is determined in the MS by the time taken by the fragments to reach the detector; this time is called as time of flight or TOF: Fragments with smaller mass reach the detector in much less time compared to fragments of with higher mass. The value of TOF is a very good indicator of the mass of the fragment; their size i.e whether such fragments contain one or more amino acids and the nature of the amino acids that these fragments are made of as shown in the table below (Table 1) (Figure 5).
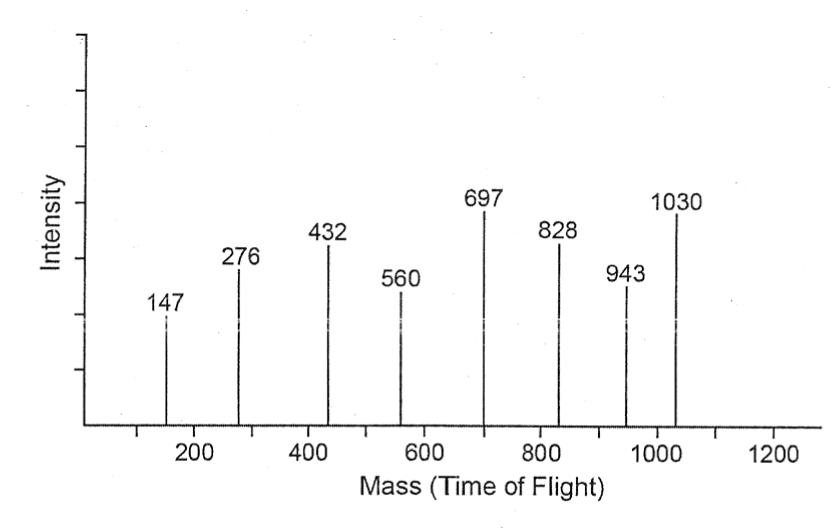
Figure 5. Characterization and identification of peptides by MS (Based after Mishra 2010 with permission of John Wiley and Sons)
Proteomics as basis for Differentiation: Differentiation is the result of expression of different set of genes producing different proteins in an organism as seen below in the development of moth from larvae (Figure 6 and 7).

Figure 6. Larvae and adult stages of moth (courtesy of Professor Richard Vogt)
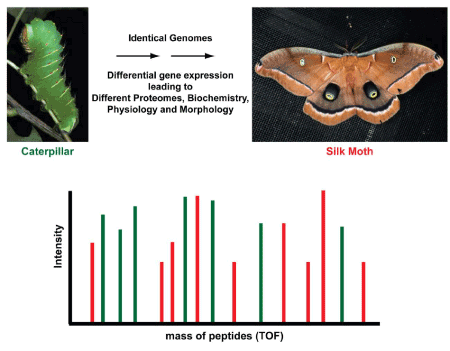
Figure 7. Diffrential expression of proteins in larvae (green) and adult moth (After Mishra 2010 with permission of John Wiley & Sons)
Complexity of organism based on Interactions of Proteins (Interactomics): It has been known for a long time that the amount of DNA in an organism does not determine its complexity as shown in (Table 1). However, what has become obvious now that the complexity of an organism is on the number of interactions of protein in an organism as shown below in Table 3. The data in this table show that even rice genome size is the largest among the organisms listed in this table it is far less complex than human which has a genome size much smaller than rice but has far more number of protein –protein interactions. Also, the data in this table show that complexity of organism is related to number of protein-protein interaction in any particular organism (Table 3,4).
Table 3.Estimated number of protein interaction in different organisms
Organism (#genes) |
Number of protein interaction |
Rice |
37000 |
Human (23,000) |
650,000* |
Worm (19,000) |
~200,000* |
Fruit fly (14,000) |
65,000* |
Table 4. List of Proteins approved by FDA as *biomarkers of human diseases.
Marker |
Disease |
CEA |
Malignant pleural effusion |
Her/neu |
Stage IV breast cancer |
Bladder Tumor Antigen |
Urothelial cell carcinoma |
Thyro-globulin |
Thyroid cancer metastasis |
Alpha fetoprotein |
Hepatocellular carcinoma |
PSA |
Prostate cancer |
CA 125 |
Non-small cell cancer |
CA 19.9 |
Pancreatic cancer |
CA 15.3 |
Breast cancer |
Leptin, Prolactin Osteopontin and Insulin, Like growth factor II |
Ovarian cancer |
Troponin |
Myocardial infraction |
B-type natriuretic peptide |
Congestive heart failure |
(Data in Table 4 Based by Polanski and Leigh Anderson (2006))
* Data based on statistical estimates by Stumpf and others (2008).
Diseasome concept: Understanding of Genomics and Proteomics has elucidated the relation among Diseases and Genes and Proteins and has led to the establishment of the concept of Diseasomes by Marc Vidal (Goh et. Al 2007) [14] at Harvard University. It depicts the relationship as network between disease genome (gene) and diseases phenome (genetic disorder). Diseasomes show how a disease is controlled by one gene and one protein or by several genes and several proteins. This is shown graphically in the diagrams below (Figure 8).
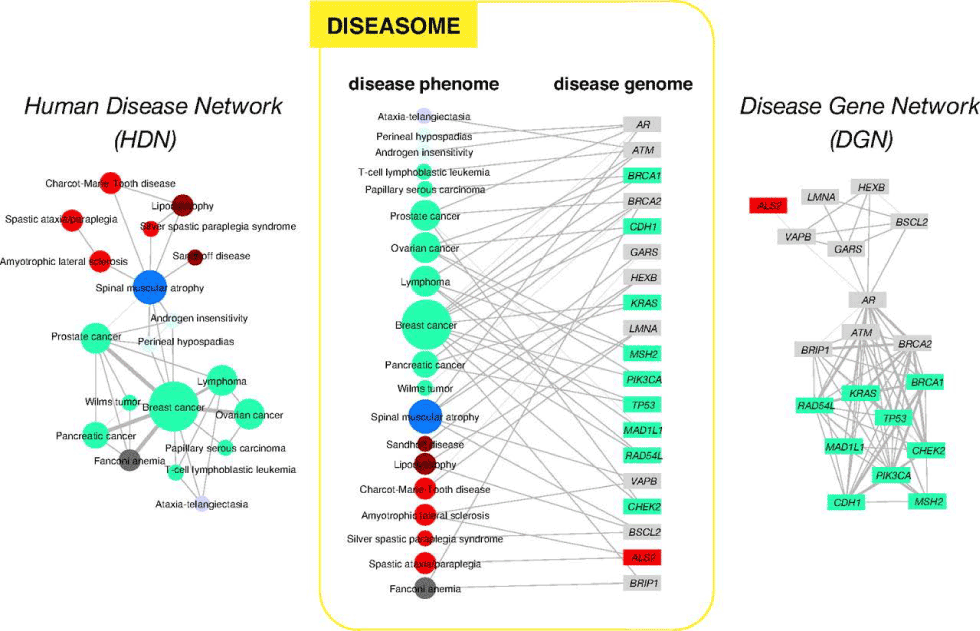
Figure 8. Network of human diseases and genes (After Goh et al. 2007 with the permission of Proceedings of the National Academy of Sciences USA)
The fact that a defective protein can cause a disease, proteins specific for a disease can be used as a marker of a disease. Such proteins are called biomarker for that disease. Identification of such biomarker can be used to screen an individual person and this can be useful in detection and cure for a disease. PSA or Prostrate Serum Antigen has been useful in monitoring an individual before the development of prostate cancer. A list of some of the biomarkers is included.
The lack of confidence in using a particular single protein as biomarker of a disease has led to the development of a panel of proteins as biomarker instead of a single protein for certain diseases.
It is shown that an increase in a combination four proteins such as leptin, prolactin, osteopontin and insulin like growth factor II taken together serves as good indicator of ovarian cancer.
None of these proteins by themselves when occurring singly in increased amount can serve as biomarkers.
Advances in Genomics and Proteomics open the possibility of personalized medicine which will use a particular drug(s) based on the genetic profile of the individuals to treat a disease: This is unlike the current approach in medicine - which is the approach of One size fits all. For example, current Cancer treatment
always involves Surgery, Radiation and Chemotherapy.
Better or specific treatment of certain diseases that may evolve into personalized medical treatment:
Cancer: Now specific drugs for cancer treatment are being developed. Some of these specific drugs include
Specific Drugs - Tamoxifen, Gleevec, Herceptin, Aromitase inhibitors Erbitux Vectibix.
In addition, Genomics & Proteomics can predict better drug response. Cancer patients metabolizes Tamoxifen in Different way for example Tamoxifen can be converted in several steps into Endoxifen with cancer fighting ability. The convertase is encoded by 2D6 gene in Human, thus patients with a mutation in 2D6 gene cannot benefit from Tamoxifen. It has been shown now that Geftinib, an inhibitor of tyrosine kinase EGFR is more effective in lung cancer in Asians. Herceptin is effective only in patients with HER-2 Mutation
Cardiovascular diseases: Understanding the genetic makeup of patients has led into prescription of doses of
Warfarin, a blood thinner.
Obesity: Obesit2021 Copyright OAT. All rights reservs of Myocytes: Brown cells which burn Fat produces 2 Zn finger proteins PRDM / C-CRB beta and white cells which controls the storage of fat.
Design of Drugs: Study of Genomic, Proteomic, Structural Biology (X-Ray, NMR) and Bioinformatics has been utilized in the design of drugs.
Metabolomics: This branch of Omics describes the role of metabolites or nutrients on the control of expression of genes in any organism. There are different sources of metabolites: such as first the metabolites produced by the organism in course of metabolic activity for example during the break down of phenylalanine in human homogenistic acid is produced which if not further metabolized lead to cause alcapatonuria, a mental disorder in which urine turns black. Second, our nutrients such as phenylalanine can cause mental disorder in certain individuals lacking or possessing a defective gene. That is the reason all newly born kids are screened for phenylketonuria and if found positive for the disease are restricted in intake of certain amino acids in their diets up to the age of 8-10 years to prevent damage to brain development in early stage of development.
Third, the metabolite that can influence our genes is the chemical produced by the microorganisms which reside in our body i.e. the activity of metagenomes.
Among the different omics we experience the effect of metabolomics more readily as we are aware that certain nutrients can cause health problem for example intake of Gluten can cause serious problems in certain individuals same is known for the intake of sugar for person suffering from diabetes or intake of milk with lactose in certain other individuals. Above all metabolic studies are much easier to carry out in human as there are only about 10000 metabolites where as there are 23000 genes and over 100000 proteins.
Epigenomics: The term epigenetics was coined by Conrad Waddington in 1912 to include changes in phenotype without any change in genotype.
Waddington was interested in embryogenesis and has found that drosophila larvae when reared at higher temperature developed into wingless flies: However, these wingless flies produced normal progeny with wings when their larvae were reared at normal temperature. These results lead him to conclude the effect of environment in the expression of genes during embryogenesis. Later he found that environmental cue can manipulate the expression of genes even in the adult life of an organism.
The landmark experiment in epigenetics was performed by Jirtle and Waterland [15] at the Duke University. In their experiment, they showed that expression of Agouti gene in mice can be silenced in the pups of agouti moms by changes in the diet of the pregnant agouti mice. Agouti gene gives the yellow color to mice and also makes them obese and prone to cancer and diabetes. Jirtle and Waterland found that agouti females produced normal puppies which were non-obese or thin and brown in color when the female agouti were fed with diets rich in supplements like vitamin 12, folic acid, choline and betaine before, during after pregnancy. These pups however produced agouti progeny on regular diet without any supplement suggesting that their genotype has not changed but just silenced. Later on, it was shown these supplements were rich providers of methyl groups and caused the gene silencing by heavy methylation of Agouti gene.
Similar experiments have been carried out in rats and even in human to examine the effect of gene silencing via methylation. These experiments were carried out by Manley and Szyf from McGill university. Such experiments examined the social behaviors in rats. Two different kinds of rat mothers were used in such studies one attentive mother which licked the pups patiently after birth like loving a mother; these pups grew to be very calm and handled the stressful situation very well and the other kind of inattentive mother which neglected the pups and did not lick them at all; in the absence of tactile experience these pups grew up to be very nervous skitter and preferred to isolate themselves in a dark corner under stress.
On further investigation, it was found that pups with tactile experience from loving mom has well developed hippocampus releasing very little of the stress hormone, cortisol with very low level of methylation. Whereas the other pups lacking tactile experience from mom with nervous disposition had very poorly developed hippocampus with lots of DNA methylation in the hippocampal cells. Based on these results Manley and Szyf suggested the role of methylation underlying the social behavior of rats. This conclusion was further enforced by the finding that nervous pups of inattentive mother became calm like the pups of attentive mother when infusion of their brains
With trichostatin A, a drug which is known to remove methyl groups. It seems pertinent to mention that trichostatin A is very much chemically similar to drug valproate which is used as mood stabilizer for certain human patients by physicians.
Later, Manley and his collaborators tried to extend their work to human even though such experimentation in human is not only difficult but impossible. They selected two groups of human subjects, one which has good relation with their mothers and another which has bad relation with their mother. These two groups were kind of equivalent to rat pups of attentive and inattentive mother. They compared the MRI (magnetic resonance image) scan of the brains of the two groups of individuals and found that the brain structures of the two groups of human individuals were significantly different. Results of these kinds of studies suggested that good mothering and bad mothering can cause difference in the brain structure in human as well leading to control of behavior.
Ordinarily such epigenetic changes due to methylation of DNA is wiped out during the formation eggs and sperms. However, at times such epigenetic differences can be passed to several generations. Epigenetic changes are brought by methylation DNA in chromosomes. It has been known that X and Y chromosomes are differently imprinted by methylation of certain epigenetic sites in these chromosomes which lead to different kinds of syndromes in human including mental retardations. Methylation of certain genes can cause cancer in human. In view of this fact FDA in the USA has approved the use of 5’azacytidine for the treat of certain cancer patients. Also, Folic acid has been approved by FDA as supplement in diet of pregnant women for the prevention of spina-bifida.
Future of the sciences of Omics: In addition to understanding of several complex diseases and design of drugs advances in Science of Omics Particularly Genomics & Proteomics will lead to a great Prospect for personalized Medicine. Some of these are summarized below.
1. Future of proteomics lies in its ability to design better drugs for human diseases. It is believed that proteomics will provides better means for the diagnosis, treatment and methods to monitor the efficacy of a drug treatment. It is expected to design better diagnostics and smart drugs by the use of the biomarkers revealed from the proteomic analysis of normal and diseased person.
Identification of protein biomarkers, their modification and altered metabolic path way by comparison of the proteomes of normal cell and cell from a diseased person will be used to design smart drugs.
Proteomics is expected to reduce the cost by increasing the number of target proteins used for drug designed.
Knowledge of metabolic pathways and that of proteins-interactions and bioinformatics tools will be used to facilitate the development of drugs in a cost-effective manner methods in a cost-effective manner. This will be aided by use of combinatorial chemistry and the library of chemicals available on the database. Since most protein work after phosphorylation by protein kinase their action can be controlled by kinase inhibitor; study of kinase inhibitor has become a field itself.
Proteomics will also lead to the discovery of new proteins as drugs. This possibility is shown by the recent studies of Mark Tuszynski [16] and his group (2001, 2011) at the University of California, San Diego. There the researchers have shown that the symptoms of Alzheimer disease such as memory loss, brain cell degeneration and cognitive impairment can be overcome by injection of a brain derived neurotrophic factor (BDNF), a protein into mice, rhesus monkey and other model animals. The BDNF protein is usually produced in the brain of normal animals but its production by cortex is missing in animals with disease. In addition to the development of drugs, proteomics is expected to help in the production of vaccine in the future.
Scientists under the leadership of professor Seth Grant at the Welcome Trust Sanger [17-19] Institute and Edinburgh University have studied human brain samples to isolate a set of proteins that account for over 130 brain diseases. The brain, the most complex organ in our body contains millions of nerve cells connected by billions of synapses. Each synapse has a set of proteins which bind together to build a molecular machine called the postsynaptic density (PSD) [20]. Professor Seth Grant and his team have extracted the PSDs from synapses of patients undergoing brain surgery and discovered their molecular components using methods of proteomics. They found 1461 proteins in human synapses, each one encoded by a different gene.
These diseases include common debilitating diseases such as Alzheimer's disease, Parkinson's disease and other neurodegenerative disorders as well as epilepsies and childhood developmental diseases including forms of autism and learning disability.
Our dilemma: There is a certain philosophical under pinning of the understanding of the science of Omics. We for centuries thought that our fate is in the stars then with the coming of DNA we started thinking that our fate is in genes and that throw out the concept of free will. However, with the understanding of epigenetics we come to think that our fate is not completely sealed in genes but there are certain windows and expression of genes can be manipulated by controlling methylation patters of genes; this after all provided some room of the idea of free will as perpetuated by our philosophers and social thinkers. Omics provides a possibility of a link from genetic engineering to social engineering causing many ethical dilemmas.
Some of our dilemmas as a result of advances in the science of Omics such as possibility of redesigning our genome by genetic engineering or controlling our behavior or social engineering by altering the methylation pattern are very much the same of that of the girl in front of mirror as depicted in the famous painting by Picasso in 1930s.
This article is dedicated to the memory of my Ph. D Supervisor and mentor Professor Steven F. H. Threlkeld of McMaster University. Author wishes to thank Professor Michael Felder, Marty Unger and Karen Unger for their support during this work.
- Lederberg J (2001) Ome sweet ome - a genealogical treasury of words. Scientist 15, 8 Anfinsin C.B. 1973 Principles that govern the folding of protein chains. Science 181: 223.
- Beadle GW, Tatum EL (1941) Genetic Control of Biochemical Reactions in Neurospora. Proceedings of the National Academy of Sciences of the United States of America 27: 499-506.
- Merrifield RB (1963) Solid Phase Peptide Synthesis I The Synthesis of a Tetrapeptide. J Am Chem Sci 85: 2149–2154.
- Gunter B (1999) Nobel Lecture: Protein Targeting. https://www.nobelprize.org/nobel_prizes/medicine/laureates/1999/blobel-lecture.html
- Gorrod AE (1909) Inborn Error of metabolism Oxford, UK, Oxford University Press.
- Yanofsky C (2005) Using studies on Tryptophan metabolism to answer basic biological questions. J Biol Chem 278: 19859-10878.
- Edman P (1950) Methods for amino acids sequence in peptides. Acta Chemica Scandinavia 4: 283-284.
- O’Farrell PH (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250: 4007-4021.
- Klose J (1975) Protein mapping by combined electrophoresis and eletro focusing in mouse tissue. Humangenetik 26: 231-243.
- Mishra NC (2010) Introduction to Proteomics: Principles and Application. John Wiley and Sons, New York 1-200.
- Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM (1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246: 64-71. [Crossref]
- Fenn JB (2002) Eletrospray wing for molecular elephants. Lex Prix, Nobel.
- H Waki, K Tanaka, Y Ido (1988) Protein and polymer analysis uo to m/z100.000by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrum 2: 151-153.
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL (2007) The human disease network. Proc Natl Acad Sci USA 104: 8685-8690
- Waterland RA, Jirtle JL (2003) Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Molecular and Cell Biology 23: 293-300.
- Lu, Mark, a Blesch and Mark Tuszynski (2001) IGF-I Gene Delivery Promotes Corticospinal Neuronal Survival but Not Regeneration After Adult CNS Injury. Journ comparative Neurology 436: 456-470
- Sanger F (1952) The arrangement of amino acids in proteins. Adv Protein Chem 7: 1-28
- Sanger F (1958) The chemistry of insulin. Nobel lecture 7: 544-556
- Sanger FS, Nicklen, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467
- Bayes A (2011) Characterization of the proteome, diseases and evolution of post synaptic density. Nature Neuroscience 14: 19-21








