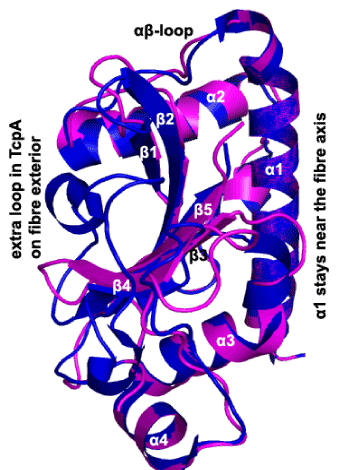Summary
The three dimensional structures of four type IVbpilin globular domains in Vibrio cholerae (TcpA), Salmonella entericserovarTyphi, enteropathogenicEscherichia coli (EPEC) and enterotoxigenicEscherichia coli (ETEC) reported earlier are compared.By superimposing in stereo, it is seen that three of the four pilins possess five super imposable β-strands, four nearly super imposable α-helices and a disulphide bond, while the bundle-forming pilin (BfpA) domain of EPEC in 1zwt does not superpose well, exhibiting a different strand topology, quite different locations for its four helices and several hydrophobic cores instead of a larger one.As a result, 1zwt lacks the extensive side chain-side chain interactions of α1 with almost parallel, three central β-strands observed in all other type IV pilins.Further, a complete pilus built using 1zwt cannot form the α2-α4 lateral interactions between consecutive subunits within the left-handed three-start helices forming the fiber as suggested for TcpA, based on the crystal structure of its globular domain.As both BfpA and TcpApili possess nearly identical fiber diffraction parameters, we think they should share a common fiber formation mode.Our alternative model for BfpA is based on the V. choleraepilin crystal structure.
Keywords
Adhesion; Fiber-Forming Protein; Type IvbPilins; Protein Folds
Virtually all gram-negative bacteria including several human pathogens contain type IV pili, which are assembled from thousands of identical subunits of a single protein called pilin plus a few copies of pilus associated proteins [1,2].In addition to host cell adhesion [3], these pili help in motility on solid surfaces [4], microcolony formation via auto-aggregation [5], bacteriophage adsorption [6]and are in general important in the virulence [5,7]of these pathogens.
Type IV pili are usually 1-4 µm long, 50-80Å diameter, flexible filaments.Type IV pilins have about 140-200 amino acids in their protein sequences, share N-terminal sequence similarity including an almost invariant glutamate at residue 5, and a single disulphide linkage near the C-terminus [1].Further, subdivision of the type IV pili into types IVa and IVb was made on the basis of endoproteolytic cleavage characteristics, length of leader sequences and the N-terminal residue of the mature pilin (1).Among human pathogens, N. gonorrhoeae, N. meningitidisand P. aeruginosapili belong to the type IVa subclass which belong to a wide range of bacteria with diverse tissue and organ specificities, while the remaining pathogens that colonize in the human intestine, including toxin-coregulatedpilus from V. cholerae, PilS from S. typhi, bundle forming pili from EPEC and colonization factor antigen III (CFA/III or CofA) or longus from ETECbelong to the type IVb subclass.
It is not easy to crystallize these pilins.The first success came with the N. gonorrhoeaepilin, crystallized in the presence of a detergent, and its X-ray structure [8].The N-terminal 52 residues form a long S-shaped α helix, its latter half associated with a globular domain.The N-terminal helix was predicted to mediate fiber formation via hydrophobic association.Next, the structures of the globular C-terminal domains of two closely related P. aeruginosapilins were solved by crystallography [9]and NMR [10]by removing the protruding half of the N-terminal α helix.Despite low protein sequence similarity, the globular domains of these P. aeruginosapilins were found to be similar in fold and disulphide bond arrangement to the full length N. gonorrhoeaepilin[8,9].Based on the N. gonorrhoeaepilin crystal structure, mutational data and secondary structure predictions etc. a theoretical model [11]was constructed for V. choleraeTcpA before the 1.3Å crystal structure of this type IVb globular domain became available [12].X-ray crystal structures of truncated TcpA and the full length P. aeruginosapilin, as well as cryo-electron microscopy analyses of their pili to suggest fiber formation from those pilins, were published together [12].
The truncated TcpA of V. cholerae is the first crystal structure of a type IVbpilin[12], though the full TcpA could not be crystallized.For two more truncated type IVbpilins, PilS and BfpA, their solution NMR structures were reported next [13,14].The third crystal structure of a full length type IVapilin from D. nodosus and another globular domain from F. tularenis were solved [15].Recently, the 1.26Å crystal structure of another truncated type IVbpilin, CFA/III or CofA was reported [16], enabling us to compare these enteric pilin structures for their similarities and differences.Comparison of experimentally obtained three dimensional structures of proteins with similar or identical function is a very powerful technique.From our comparison of these four experimentally determined structures, we conclude that the NMR structure of BfpA globular domain [14]may be a result of a folding that is biologically not relevant, hence we have constructed an alternative model by homology.
Materials and Methods
Sequence alignment
An initial multiple protein sequence alignment was carried out with the mature pilin sequences in TcpA, CofA and BfpA using ClustalW with default parameters before examining the experimental structures for the latter two.This alignment did show the 10-residue extra loop in CofA relative to TcpA occurring between the two helices α1 and α2 (Figure 1, 2) verified by the recent crystal structure 3s0t [16].However, the alignment was not judged to be satisfactory near the C-termini.From the TcpA crystal structure [12], we concluded that the easiest way to retain the same architecture for BfpA having 19 residues less than TcpA is by a much smaller loop between β3 and β4.Further, the two Cys residues in BfpA were manually aligned with those in TcpA and CofA, assuming a conserved position of the disulphide bond and the two associated helices α3 and α4.
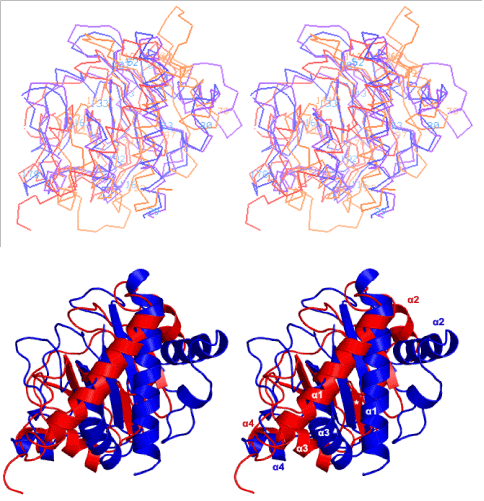
Figure 1:(A) Four type IVbpilin globular domains as Cα models are superimposed in stereo. High-resolution crystal structures of TcpA (blue) and CofA (violet); NMR structures of PilS (orange) and BfpA (red), are superposed pair wise using the β-strands forming the central β-sheets. Residues at the start of secondary structure elements are numbered in light blue (TcpA) and orange (PilS). It is seen that TcpA, CofA and PilS share the same topology in their central 5 β-strands and the relative dispositions of four conserved helices α1 through α4 relative to their central β-sheets, but the topology of the BfpA NMR structure (red) is different. The strange topology of BfpA region 60–90 prevents its α1 from coming closer to its central β-sheet and this α1 is roughly at an angle of 60o relative to its β-strands; they are roughly parallel in the others. (B) Only TcpA (blue) and the mixed sheet NMR structure of BfpA (red) are superposed as ribbons, with α helices in both marked. The odd β-strand is marked with a star (*) in white.

Figure 2. Sequence alignment of TcpA (PDB: 1oqv), CofA (PDB: 3s0t), BfpA (PDB: 1zwt) and PilS (PDB: 1q5f). Secondary structures in the two high-resolution crystal structures in 1oqv and 3s0t, and the two NMR structures in 1zwt and 1q5f are given above each sequence. For BfpA, a second line of secondary structure is for our predicted model based on 1oqv. Additional helices such as α2’, α3’ in 1oqv, α1’ in 1zwt and β-strands in 1q5f are so named to maintain a uniform nomenclature for the important helices α1 to α4 and strands β1 to β5 among the four sequences.
Superposition of type IVb globular domains
The coordinate files for four type IVbpilin globular domain structures were obtained for TcpA (PDB: 1oqv), CofA (PDB: 3s0t), BfpA (PDB: 1zwt) and PilS (PDB: 1q5f).For the two crystal structures, only subunit A was chosen, and the first among the several within the files for the two NMR structures.The protein structures were displayed in stereo with various options using the program InsightII[17].Backbone atoms in regions 102-110, 117-149, 174-178 and 200-208 of 3s0t were superposed pair wise with regions 92-100, 105-137, 162-166 and 191-199 of 1oqv respectively.With this superposition in place, backbone atoms in regions 91-96 and 103-109 of 1zwt only (residue nos. increased by 22 to give those in the mature pilin) could be superposed with regions 93-98 and 107-113 of 1oqv, as 1zwt possesses a different topology in its central β-strands [14].Backbone atoms in regions 98-103, 113-119, 141-145 and 173-180 of 1q5f were superposed with regions 93-98, 107-113, 133-137 and 192-199 of 1oqv respectively.
New BfpA model construction
Using a pair wise alignment between TcpA and BfpA sequences within the Homology module of InsightII[17], close to the one indicated in Figure 2, coordinates for the structurally conserved regions in the BfpA globular domain were obtained.TcpA was chosen as the template rather than CofA or PilS, as in our multiple sequence alignment, the overall similarity was greater with TcpA.Six loop regions, α1-α2, α2-β1, β1-β2, α3-β3, β3-β4, β4-α4 constituting variable regions, were built manually within InsightII, and coordinates merged with the remainder of the protein.These six regions were energy refined within InsightII using the program Discover first, holding the remainder of the protein fixed.After the regularization of these manually built variable regions, the side chains of the structurally conserved regions were allowed to move during further energy minimization cycles until acceptable stereochemistry, bond lengths (1 bond deviating > 4σ) and bond angles (10 angles deviating > 4σ) were obtained, and 92% of the residues were in Ramachandran preferred / allowed regions.This new BfpA model replaces our older model (PDB: 1qt2) which was based partly on the N. gonorrhoeaepilin.
Results and Discussion
Sequence similarities and globular domain structures compared
The TcpA-CofA pair has 64/199 or 32.2% sequence identity overall; omitting the 30 N-terminal residues, they share 41/169 or 24.3% sequence identity in their globular domains.In addition, there are highly similar residues in 44/199 positions or 41/169 positions between the globular domains.This pair has an RMSD of 1.4 Å considering all the Cα atom pairs [16].The superposed regions involving 224 atom pairs used for TcpA-CofA in Fig. 1 have an RMSD of 0.98Å.
The TcpA-BfpA pair has 40/180 or 22.2% sequence identity overall; omitting the 30 N-terminal residues, they share 28/150 or 18.6% sequence identity in their globular domains.Also, there are highly similar residues in 38/180 positions or 30/150 positions between the globular domains.Superposed regions involving only 52 atom pairs used for Fig. 1 have an RMSD of 1.14Å for TcpA-BfpA, other three strands roughly superposing with a reversed chain direction for Cα atoms.
The TcpA-PilS pair has 20/181 or 11.0% sequence identity overall; omitting the 30 N-terminal residues, they share 17/151 or 11.3% sequence identity in their globular domains.There are highly similar residues in 38/181 positions or 31/151 positions between the globular domains.The superposed regions involving 104 atom pairs have an RMSD of 1.49Å for TcpA-PilS.
The enteric pilins have not been previously compared in the various papers by superposition in stereo [14,16]though ribbon diagrams for the globular domains were provided.In order to provide an overall comparison of the three-dimensional structures of the different pilins, it is most insightful to superimpose the central β-strands alone rather than other portions of the structures and this is the procedure we have followed (e.g., see Fig. 1A).
Our superposition in Figure 1A shows that TcpA (blue), CofA (violet) and PilS (orange) have practically super imposable five β-strands, corresponding to β1-β2-β5-β3-β4 of TcpA and CofA, forming purely anti-parallel β-sheets that are curved as if on a barrel surface.Helices α1-α4 got roughly superimposed among these three protein structures even though their coordinates were not used in the superposition.The BfpA (red) structure 1zwt has a mixed β-sheet as its region 81-83 is parallel to an adjacent strand; hence it has a different topology.This same region prevents its N-terminal α-helix from coming closer to its β-sheet to form a sizeable hydrophobic core as observed in the three other typeIVbpilins (Fig. 1).TcpA and CofA are especially close to each other, the extra element in the latter being a loop between α1 and α2 [16].In 1oqv and 3s0t, there are some main chain-main chain hydrogen bonds between β4 and the β3-β4 loop seen at the extreme left in Fig. 1, though they do not define a β-sheet.PilS (orange) differs from TcpA and CofA in two main regions – following α2, there are two additional β-strands which form part of the 5-strand β-sheet extending it to the right, and the β3-β4 loop is much shorter in PilS.These differences can also be seen in our alignment shown in Figure 2.
BfpA model based on TcpA crystal structure
In the model proposed here,BfpA has 3 residues (135-137) compared to 23 residues in TcpA (139-161) in the β3-β4 loop (Figure 2, 3).This is the major difference between the structures.TcpA and PilS also have a similar difference.This is reasonable as the number of residues in PilS and BfpA are very close.However, for the so-called αβ-loop region, BfpA is closer to TcpA in structure as well as total number of residues; PilS has a few more residues in this region forming its additional two β-strands (Figure 2).In the β4-α4 loop on the other hand, PilS has a smaller number of residues compared to TcpA and BfpA (Figs. 1, 2).
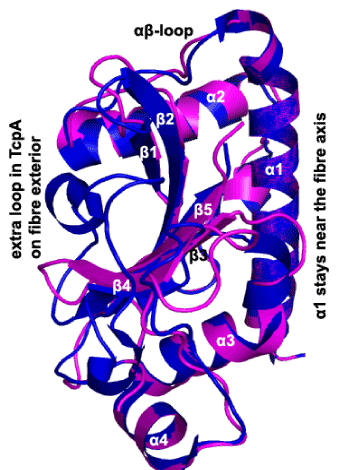
Figure 3:Ribbon diagram of our BfpA globular domain model (magenta), superposed on the TcpA globular domain (blue), with various secondary structure elements labeled. BfpA has a 3-residue (135-137) loop between β3 and β4 compared to the much longer loop shown for TcpA. This longer loop extends both above β4 in this view (next to the label provided, to the left) and between β4 and α4 where no corresponding magenta segment can be seen.
The 35 side chains participating in the hydrophobic core in the model proposed here are displayed in Figure 4.Out of 35 such side-chains participating in the hydrophobic core in this model, only S33A, P95A, A165V and K174T mutations are observed; the remaining 31 are conserved despite variations observed among BfpA strains [18].In this model, side chains of S33, A36, I37 and A40 participate in the main hydrophobic core, but they do not in 1zwt.The NMR structure [14] described several two hydrophobic cores containing 8 + 7 or 15 side chains (Val143 in that list should be Val133) instead of a much more extensive core as in our model (Fig. 4).However, there is a third cluster of hydrophobic side chains in 1zwt on the first face comprising of I47, L50, Y51, Y57, F 87, L91, Y104, L106 and L106 not mentioned by Ramboarina et al. [14].There is a fourth cluster of hydrophobic side chains comprising of L65, P72, Y75 Y105 residues on the opposite face, also not mentioned [14].The proposed model has a small hydrophobic patch on the ‘fiber exterior’ side of the β-sheet, comprising residues Y105, T107, V175 and Y177.Extremely variable regions like 58-63, 97-101, 137-155, 167-169 [18]are expected to be exposed in the fiber exterior in the model proposed (Figure 5).
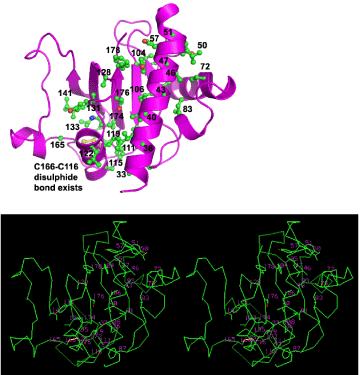
Figure 4:One continuous hydrophobic core of our BfpA model. (A) The model is shown as a ribbon in magenta with numbered side chains comprising the core in ball-and-stick. (B) The same side chains composing the hydrophobic core embedded in Cα backbone for our model, in stereo. Out of 35 side-chains forming the core, 31 are fully conserved among 8 BfpA sequences and 4 show conservative mutations. α1 is roughly parallel to 3 β-strands. Side chains from a loop after α2 and some from α3 are also seen to participate in this hydrophobic core.

Figure 5:Displaying two adjacent subunit globular domains in BfpA as proposed for TcpA as left-handed three start helices (Fig. 4 of ref. 12), emphasizing α2-α4 interactions between these neighboring subunits comprising the fiber; β-strands are marked by numbers only. Side chains at the interface and their sequences observed in 8 different BfpA strains are displayed.
Pilin biogenesis
It was noticed that expression of D. nodosus, M. bovis and N. gonorrhoeaepilin genes in P. aeruginosa results in the formation of pili.Thus the basic machinery involved in the biogenesis of the type IVapili appear to be conserved [1].However, full length TcpA expressed in EPEC failed to form fibers since prepilins could not be completely converted to pilins by the EPEC prepilin peptidases [19].This insufficient conversion could be simply due to the difference in the N-terminal amino acid (Met vs. Leu) between the two pilins and does not justify the reported divergence in fold [14].CofA has Met like TcpA and is synthesized and processed by V. cholerae, but the TCP assembly apparatus cannot make filaments from CofA (16).Yet, the topology of TcpA and CofA are identical.
Arguments in support of homology model
It is very surprising that PilS having 21.5% sequence similarity with TcpA in the globular domain still possesses the same topology, but the NMR structure of BfpA having 28.6% sequence similarity, has a different fold.The proposed BfpA model however has the topology and fold as the high resolution TcpA crystal structure.
The globular domain of BfpA has 28.6% sequence similarity with that of TcpA, whereas it is 36.4% for the CofA-TcpA pair, already demonstrated by two high resolution crystal structures [12,16] to have an RMSD of 1.4Å.A crystal structure of residues 26-181 of PilS[20]shows local differences with its NMR structure [13], with 93 Cα pairs having an RMSD of 1.87Å.The PilS crystal structure shows a longer α1 and different locations for β-1 and β0 (defined in Fig. 2) relative to the NMR structure.The PilS crystal structure shows an RMSD of 1.73Å with TcpA using only 93 Cα pairs (20), with 21.5% sequence similarity.From sequence similarity alone, it is likely that an experimental structure of BfpA will show an RMSD intermediate between 1.4Å and 1.7Å with the TcpA crystal structure [12].For sequence identity in the 25-29% range, half the models predicted using SWISS-MODEL show less than 4Å RMSD generally (www.bioinf.ac.uk), but in the case of type IVbpilins, three dimensional structure is conserved better despite sequence variation.
By sequence alignment alone, we were able to predict the 10-residue extra loop in CofA relative to TcpA before the crystal structure of CofA was published [16].This shows the power of protein sequence alignment in correctly predicting the loop when an appropriate template structure is used.
Our BfpA model resembles 1zwt in secondary structure (Fig. 2).Hence, it satisfies a major part of the NMR data related to secondary structure [14].
All type IV pilin structures, including our model, show an intimate association of side chains from α1 with those from three β strands (β1-β2-β5 in type IVb, β1-β2-β3 in type IVa).This feature is observed in all structures except in 1zwt.It is thus inconsistent with other pilin structures.
With the 181 residue PilS, both X-ray and NMR show a small β3-β4 loop (termed β5-β6 in those studies) as proposed in our model for the 180 residue BfpA.The PilS structure can be fitted in the same pilus arrangements derived for TcpA from EM data and packing arrangements [20], though a different fiber formation mode was suggested earlier [13].
Simulations favor the formation of a single hydrophobic cluster midway through a globular protein core formation process [21], as in the proposed model.But several smaller cores exist in 1zwt, as described above.By running the program Discover within InsightII, our final model of BfpA globular domain (residues 29-180) attained a total energy of -12,250.1 kcal, whereas that for the TcpA crystal structure (residues 29-199) after similar refinement was -10,249.9 kcal.With the NMR model 1zwt (residues 23-180), a total energy of -9,209.5 kcal was reached.The main source of energy difference between the proposed model and 1zwt was in the Coulomb energy, of about 3,000 kcal.Further, the CATH database (www.cathdb.info) has assigned the same TcpA-like pilin fold 3.30.1690.10 for 1oqv, 1q5f and 3s0t, but none for 1zwt.
Other reasons for the fold in 1zwt
Even if we accept that 1zwt was obtained by correctly analyzing the NMR data, there could be additional reasons why the fold observed therein is unusual.One assumption with all these truncated pilin studies is that the α1 truncation does not affect the globular domain structure; this assumption may not be true for 1zwt which deleted 22 residues.Secondly, the first two N-terminal amino acids in 1zwt are Met and Asp (residues 23, 24) respectively, whereas the BfpA sequence has Tyr-Tyr (Figure 2). This too may have pushed α1 away from the central β-sheet, as both of these amino acids are found to be hydrophobic in all type IVbpilins (Figure 2).
Fiber formation
From EM analysis, the distance between adjacent left handed helices formed by the TcpA molecules within the TPC fiber was found to be 45Å, and a rise of 22.5Å between adjacent TcpA molecules together with 60o rotation about the fiber axis [12].These dimensions obtained from the EM analysis of the BFP fibers were 44Å and 22Å respectively, i.e., nearly identical [14].We accept that the EM data of BFP fibers also supports a three-start helix [14].The N-terminal helix is agreed to be roughly parallel to the fibers in all type IV pilins.
It is shown that our BfpA model closely follows the template TcpA structure (Figure 3) except for the β3-β4 loop, which is on the exterior side of the fiber and therefore not directly involved in fiber formation [12].Since the subunit structure is proposed to be close, including the α2 and α4 regions important in lateral contacts, it immediately follows that our BfpA structure will be able to mimic TcpA in fiber formation.
It was predicted that, two classes of residues forming the core of the monomer and those involved in monomer-monomer interactions would be conserved within various strains within V. cholerae and a variation in residues on the fiber exterior would be observed [11].These predictions were verified in the TcpA crystal structure [12].The same principles would apply to BfpA of EPEC, another member of the type IVbpilins.It has been mentioned that 31 of the 35 residues forming the hydrophobic core (Fig. 4) are fully conserved among 8 EPEC strains, while the remaining 4 residues show conservative mutations.Regarding residues involved in monomer-monomer interactions, the αβ-loop region 65-77 (including α2) and region 156-165 (including α4) immediately preceding the second Cys are important for the fiber formation as suggested within each strand of left-handed three start helices [12]; they are all conserved among the BfpA strains [18](Figure 5).In the lateral subunit-subunit interactions, N67, T68, I71, Y75 may be interacting with T164, A163, A160 and P159 predicted to be important in such α2-α4 interactions are all conserved among the EPEC strains (Fig. 5).Extremely variable regions like 58-63, 97-101, 137-155, 167-169 among the strains [18] will be exposed in the fiber exterior constructed with our BfpA model, related to its epitopes.
If α2 of one molecule should interact with α4 of the adjacent molecule, the component of the α4-α2 vector should be about 22.5Å along the fiber axis for their interaction to occur [12].For example, Cα atoms of E183 and K68 in TcpA are 35.26Å apart, and their inter-atomic vector makes an angle of 50owith the α1; so the component equals 35.26 cos 50oÅ or 22.66Å.However, Cα atoms of A163 and G59 in 1zwt are 38.22Å apart, and the vector is parallel to α1 in the same structure [14], so there is no way for α2 and α4 to interact in a lateral fashion for forming any of the three-start helices.A model for pilus formation with CofA was generated using the TCP electron microscopy reconstruction [16], and the extra 10-residue insert fits perfectly within the gap between the CofA globular domains.
In summary, the biological insights emerging from our comparative study seem to be the unity in diversity in the type IVbpilins, marked by the two classes of conserved residues and variability in fiber-exposed residues among strains including the predictions about specific BfpA residues being involved in the lateral contacts, delineation of their common secondary structural elements α1-α4 and β1-β5 as shown in Fig. 2, the existence of a common mode of fiber formation among type IVbpilins characterized by the α2-α4 interaction [12], a common association of α1 with three central β-strands extending to all type IV pilins and existence of a smaller β3-β4 loop in smaller type IVbpilins (PilS, BfpA) compared to the larger two (TcpA, CofA).
Acknowledgments
Bose Institute is thanked for providing InsightII and other facilities for our study.The authors also thank Mrs. DevlinaChakravarty of our department for help in preparing some figures.The authors also declare they have no conflict of interest through this study.
2021 Copyright OAT. All rights reserv
Supplementary Data
Supplementary figures and coordinate file for the new BfpA model.
References
- Strom MS Lory S (1993) Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol 47: 565-596. [Crossref]
- Rudel T Scheurerpflug I, Meyer TF (1995) Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature 373: 357-359. [Crossref]
- Doig P, Sastry PA, Hodges RS, Lee KK, Paranchych W, et al. (1990) Inhibition of pilus-mediated adhesion of Pseudomonas aeruginosa to human buccal epithelial cells by monoclonal antibodies directed against pili. Infection and Immunity 58: 124-130.
- Bradley DE (1980) A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol 26: 146-154. [Crossref]
- Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, et al. (1998) Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280: 2114-2118.
- Waldor MK Mekalanos JJ (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272: 1910-1914. [Crossref]
- Herrington DA, Hall RH, Losonsky G, Mekalonos JJ, Taylor RK, et al. (1988) Toxin, toxin-coregulatedpili, and the toxRregulon are essential for Vibrio cholerae pathogenesis in humans. Journal of Experimental Medicine 168: 1487-1492.
- Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, et al. (1995) Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378: 32-38.
- Hazes B, Sastry PA, Hayakawa K, Read RJ, Irvin RT, et al. (2000) Crystal structure of Pseudomonas aeruginosa PAK pilin suggests main-chain dominated mode of receptor binding. Journal of Molecular Biology 299: 1005-1017.
- Keizer DW Slupsky CM, Kalisiak M, Campbell AP, Crump MP, et al. (2001) Structure of a pilin monomer from Pseudomonas aeruginosa: implications for the assembly of pili. J BiolChem 276: 24186-24193. [Crossref]
- Chattopadhyaya R, Ghose AC (2002) Model of Vibrio cholerae toxin coregulatedpilin capable of filament formation. Protein Engineering 1: 297-304.
- Craig L, Taylor RK, Pique ME, Adair BD, Arval AS, et al. (2003) Type IV Pilin Structure and Assembly : X-Ray and EM Analyses of Vibrio cholerae Toxin-CoregulatedPilus and Pseudomonas aeruginosa PAK Pilin. Molecular Cell 1: 1139-1150.
- Xu XF Tan YW, Lam L, Hackett J, Zhang M, et al. (2004) NMR structure of a type IVbpilin from Salmonella typhi and its assembly into pilus. J BiolChem 279: 31599-31605. [Crossref]
- Ramboarina S Fernandes PJ, Daniell S, Islam S, Simpson P, et al. (2005) Structure of the bundle-forming pilus from enteropathogenic Escherichia coli. J BiolChem 280: 40252-40260. [Crossref]
- Hartung S, Arval AS, Wood T, Kolappan S, Shin DS, et al. (2011) Ultrahigh Resolution and Full-length Pilin Structures with Insights for Filament Assembly, Pathogenic Functions, and Vaccine Potential. Journal of Biological Chemistry 286: 44254-44265.
- Kolappan S Roos J, Yuen AS, Pierce OM, Craig L (2012) Structural characterization of CFA/III and Longus type IVbpili from enterotoxigenic Escherichia coli. J Bacteriol 194: 2725-2735. [Crossref]
- Accelrys Inc. (2005) Insight II. San Diego, USA.
- Eric Blank T, Zhong H, Bell AL, Whittam TS, Donnenberg MS, et al. (2000) Molecular Variation among Type IV Pilin (bfpA) Genes from Diverse Enteropathogenic Escherichia coli Strains. Infection and Immunity 68: 7028-7038.
- McNamara BP,Donnenberg MS (2000) Evidence for specificity in type 4 pilus biogenesis by enteropathogenic Escherichia coli. Microbiology 146: 719-729. [Crossref]
- Balakrishna AM, Saxena AM, Mok HY, Swaminathan K (2009) Structural basis of typhoid: Salmonella Typhi type IVbpilin (PilS) and cystic fibrosis transmembrane conductance regulator interaction. Proteins: Structure, Function and Bioinformatics 77: 253-261.
- Tsai J Gerstein M, Levitt M (1997) Simulating the minimum core for hydrophobic collapse in globular proteins. Protein Sci 6: 2606-2616. [Crossref]