Abstract
Buffalo ES cells were subjected to differentiation in 15% KoSR- and 15% FBS- based spontaneous differentiation media in static suspension cultures. The embryoid bodies (EBs) so formed, were analyzed for germline-specific gene expression on day 4, 8 and 14 in order to identify the optimum differentiation strategy. Immunocytochemical analysis was performed for detection of germ lineage specific markers in EBs differentiated under the identified optimum conditions. Global DNA methylation analysis was performed to assay methylation erasure in the optimized differentiation cultures. We observed a significantly (p<0.05) increased expression of all germ lineage genes like DAZL, VASA and PLZF (PGC-markers); SYCP3, PRM2, TNP1/2 and MLH1 (Meiotic markers); BOULE and TEKT1 (Spermatocyte-markers); GDF9 and ZP2/3 (Oocyte-markers) upon spontaneous differentiation in comparison to undifferentiated ES cells. EBs collected from FBS-based medium showed significantly (p<0.05) higher expression of early germ lineage genes (DAZL, TNP1, PRM2), while those from KoSR-based medium showed greater expression of meiotic and developmental genes (SYCP3, MLH1, TEKT1, GDF9 and ZP2). Immunocytochemistry revealed expression of c-Kit, Dazl, Vasa (PGC-markers); Sycp3, Mlh1, Protamine1 (Meiotic markers); Acrosin and Haprin (Spermatocyte-makers); and Gdf9 and Zp4 (Oocyte-markers) in day 14 EBs collected from both the cultures. However, only PGC-specific markers were detected in 14 day monolayer cultures differentiated in KoSR-based media. The levels of 5-methyl-2-deoxycytidine were not significantly (p<0.05) different between EBs collected from the two media formulations. However, EBs collected from KoSR-based medium showed lower concentration of 5-Methyl-2-deoxycytidine, indicating comparatively greater methylation erasure.
Key words
buffalo embryonic stem cells, spontaneous differentiation, germ lineage, gene expression, immunocytochemistry, methylation analysis
Introduction
Embryonic stem (ES) cells possess the capability to differentiate into all the three germ layers (ectoderm, mesoderm and endoderm). This property makes them suitable for elucidation of molecular mechanisms underlying differentiation and embryonic development. Among all the cell types ES cells can differentiate into, primordial germ cells (PGCs) are the most fascinating as they represent in vitro completion of reproductive cycle of the organism from which the ES cell line is derived. Given such vital functions of germ cells for propagation of sexually reproducing organisms, their formation must be rigidly programmed which is indeed the case in lower organisms like Drosophila melanogaster, Xenopus laevis, Danio rerio and in various egg-laying species [1-3]. Unlike, the pre-determined model of germ cell specification in lower organisms, mammalian primordial germ cell formation occurs later in embryonic development under the guidance of extracellular signals and in a definite cellular environment of the indifferent gonad [4] where a subset of epiblast cells begins to express BLIMP1 and FRAGILIS genes, thereby becoming primordial germ cells. These cells suppress HOX gene expression and set themselves apart from somatic differentiation programme [5]. Under the influence of various genes like STELLA, c-KIT, DAZL and VASA, PGCs migrate through genital ridge to colonize the bipotent gonad which provides necessary microenvironment for their differentiation into gametes [6-8]. The haploidy of germ cells is achieved by the delicate process of meiosis, mediated by various proteins like Vasa, Sycp, Mlh1, Transition proteins (TNPs) and Protamines (PRMs), etc. [9,10]. Further development into mature and functional gametes occurs at puberty under the influence of mature germ cell specific proteins like Gdf9 and Zona pellucida (Zp4) (in oocyte) and Tektin, Acrosin and Haprin (in spermatocyte), etc. [11-14].
Since ES cells have the intrinsic capability to differentiate into any cell type; we investigated differentiation of buffalo ES cells into germ lineage cells under spontaneous differentiation in both monolayer adherent cultures as well as in floating cultures through formation of three-dimensional aggregates known as embryoid bodies (EB). The differentiation was assessed through detection and quantitation of RNA and protein markers associated with: i) PGC ii) Meiosis; iii) Spermatocyte; and iv) Oocyte. The understanding of germ line development is of crucial importance in farm animals, especially in poor breeding bubaline species so as to understand the genetics and biochemistry of this intricate process. The understanding is further obscured due to paucity of primary tissue (first trimester fetal gonads) and complexities of in vivo system like cellular environment and embryo manipulation [15]. The development of culture system for differentiation of species-specific embryonic stem cells into germ cells would not only abreast us with the knowledge and understanding of this process but would further help us to design the biotechnological, chemical and/or nutritional interventions for enhancing the reproductive performance of farm animals in general and bubaline species in particular. The study would bear a future application in reducing the age of sexual maturity, increasing the gamete quantity and quality, especially of oocytes whose reservoir is thought to be fixed and much lesser in buffalo as compared to cattle, development of designer gametes, transgenic animal production as well as conservation of endangered animal species.
Materials and methods
Ethical approval
The study was approved by Institute’s Animal Ethics Committee as well as by Department of Biotechnology, New Delhi, for use of animal tissues as well as for development of the embryonic stem cell lines and further subsequent studies.
Chemicals
All chemicals and media used in the present study were obtained from Sigma Aldrich (St. Louis, MO) and the plastic ware from Falcon (Paignton, UK), unless stated otherwise.
Embryonic stem cell establishment and maintenance
Three previously established and characterized buffalo ES cell lines developed from in vitro fertilization (IVF) derived blastocysts were used in the study [16]. The primary ES cell colonies were developed from mechanically isolated inner cell masses of day 7 blastocysts. The primary colonies were propagated on fetal fibroblast feeder layers and maintained in embryonic stem cell culture medium, composed of KoDMEM + 15% Knock out serum replacer (KoSR), supplemented with 2 mM L-glutamine, 5 ng/ml bFGF (basic fibroblast growth factor), 1000 U/ml rmLIF (recombinant murine lukaemia inhibitory factor), 1X nonessential amino acids and 50 mg/ml gentamicin sulphate, and incubated at 38ºC in a 5% CO2 incubator. The colonies were characterized for protein and/or RNA markers associated with pluripotency and self-renewal like OCT4, NANAOG, SOX2, STAT3, REX1, FOXD3, SSEA-1, SSEA-4, TRA-1-60, TRA-1-81, CD90, c-MYC and TELOMERASE. The cell lines were also characterized for Alkaline phosphatase activity, stable karyotype and differentiation potential to three germ layers (ectoderm, mesoderm and endoderm). The cell lines were subjected to spontaneous differentiation at passage 20-30.
Spontaneous differentiation into germ lineage cells
Differentiation in floating cultures via EB formation: ES cell colonies were subjected to differentiation by static suspension culture (SSC) strategy which was earlier shown to have more potential for inducing differentiation across all the three germ layers than the hanging drop (HD) strategy [16]. ES cell colonies were allowed to differentiate in low attachment 35 mm Petri dishes in spontaneous differentiation medium, composed of KoDMEM supplemented with 2 mM L-glutamine, 1X nonessential amino acids and 50 µg/ml gentamicin sulphate. The medium was supplemented either with 15% Knock out serum replacer (KoSR) or with 15% fetal bovine serum (FBS). Half of the culture medium was replaced with fresh medium every alternate day. EBs developed in the two media formulations were analyzed by qPCR for germ lineage gene induction at day 4, 8 and 14 of culture period. EBs were also used for immunocytochemical analysis for germ line specific protein expression as well as for Global DNA methylation to assay methylation erasure.
Differentiation in monolayer adherent cultures: ES cell colonies were cut into small clumps in 1:8 split ratios. The clumps were washed thrice with DPBS-- and treated with Trypsin-EDTA for 2-3 min to remove any attached feeder cells. This was followed by two washes in DPBS--, treatment with Accutase and repetitive pipetting, with a finely drawn glass pipette, till all the clumps dissociated into single cells. The cell pellet, obtained by centrifugation at 2000 rpm for 5 min, was seeded onto MaxGel (1:200) coated 4-well dishes for spontaneous differentiation, as described above.
qPCR analysis for germ lineage gene induction: Total RNA was extracted from 100 EBs, collected randomly from the differentiation cultures, on day 4, 8 and 14 using RNeasy RNA extraction kit (Qiagen, Germany) and employed for cDNA synthesis using SuperScript III first strand cDNA synthesis kit (Invitrogen, USA). qPCR analysis was performed for quantification of such genes associated with: i) PGC-specification and proliferation (OCT4, NANOG, DAZL and VASA); ii) Meiosis (MLH1, SYCP3, TNP1/2 and PRM2; iii) Spermatocyte (BOULE and TEKT1); and iv) Oocyte (GDF9 and ZP2 and 3). The primers and their annealing temperatures have been previously mentioned [17]. The optimized qRT-PCR reactions contained 2 μL (100ng) cDNA template, 10 μL SYBR Green PCR Master Mix Buffer (2X), and 10 pmol each of forward and reverse primers in a total volume of 20 μL. The reactions were performed on CFX96 instrument (BioRad Hercules, CA, USA). GAPDH and β-ACTIN were used as the endogenous controls. The thermal cycling parameters were: initial denaturation at 95°C for 10 min; followed by 40 PCR cycles (denaturation: 95°C for 30s, annealing at X˚C (respective for each gene as provided already; [17]. for 30s, and extension at 72°C for 30s. Following amplification, a dissociation protocol was performed to provide evidence for a single reaction product. A no-template control (NTC) of nuclease-free water was included in each run and all PCR reactions were performed in triplicates. The PCR efficiencies (E) were evaluated by a 2-fold dilution series of cDNAs (1 - 1:64 dilution) for each pair of primers including the reference genes (GAPDH and β-ACTIN) using a cDNA pool, obtained from all the EBs, to ensure that no inhibitory component was present in the samples. A graph of threshold cycle (Ct) versus relative log2 copy number of the calibration sample from the dilution series was produced and the reaction efficiency was determined for each primer set by using the slope of this graph (E=2 (-1/slope)). The percent efficiency of each primer pair was found to be higher than 98% and no inhibitory effect was observed in the samples. The expression of each target gene was normalized against the geometric mean of corresponding threshold cycles of the endogenous controls (GAPDH and β-ACTIN). The relative expressions of germ lineage associated genes were calculated using 2-∆∆Ct method, taking the corresponding values for undifferentiated ES cells as the calibrator or control values [18].
Immunocytochemical analysis for germ lineage proteins: A total of 300 EBs, collected randomly from both the differentiation cultures at day 14, were subjected to immunodetection of germ lineage markers; like c-Kit, Dazl, Vasa (PGC-specific proteins), Mlh1, Sycp3, Protamine1 (Meiotic proteins), Acrosin and Haprin (spermatocyte-specific proteins) and Gdf9 and Zp4 (oocyte-specific proteins). Immunocytochemical analysis was also performed on day 14 monolayer differentiated cultures, as per the already discussed protocol [17,19]. All the primary as well as secondary antibodies have been detailed previously [17]. The primary antibodies were diluted 1: 100 in blocking buffer (4% normal goat serum in DPBS) except for Dazl which was diluted at 1:50 ratio. The testicular and ovarian tissue sections were used as positive controls while undifferentiated buffalo ES cells were used as the negative controls. EBs were washed twice in DPBS, followed by fixation in 4% para formaldehyde (PFA) for 20 min. After two washes in DPBS, the EBs were treated with 0.1% Triton X-100 in DPBS for 15 min, followed by two washes again with DPBS and incubation for 1h in blocking solution at room temperature. The colonies were exposed to primary antibodies at 4°C overnight. After three thorough washings with DPBS, the EBs were incubated for 1h at room temperature with the appropriate class specific secondary antibody, labeled either with fluorescein isothiocyanate (FITC) or phycoerythrin (PE). After washing thrice with DPBS, EBs were put individually on a clean grease free microscopic glass slide with a narrowly drawn glass pipette and overlaid with Prolong Gold Antifade reagent with DAPI (4-6-Diamidino-2-phenylindole) (Life technologies, USA) to stain the nuclei. After 5-10 min, the colonies were covered with clean grease free microscopic cover slips followed by visualization under a fluorescence microscope (Diaphot; Nikon, Tokyo, Japan). The similar protocol was followed for immunostaining of 45 different monolayer adherent cultures differentiated for a period of 14 days.
Global DNA methylation analysis: Global DNA methylation analysis of undifferentiated control ES cell colonies as well as of day 14 EBs was performed by employing Global DNA Methylation ELISA Kit (Cell Biolabs, USA), following the previously discussed protocol [17,19]. Genomic DNA (gDNA) was extracted by NucleoSpin Tissue DNA purification kit (Macherey-Nagel, Germany), following the manufacturer’s instructions. Extracted gDNA samples were digested to nucleosides by treatment with nuclease P1 and alkaline phosphatase, as described [17]. Reaction mixture was centrifuged at 6000g for 5 min and supernatant, containing individual nucleosides, was used for 5MedCyd ELISA assay. 50 μl of sample DNA or 5MedCyd standard was added to the wells of the 5MedCyd-DNA conjugate coated plate and incubated at room temperature for 10 min on an orbital shaker. This was followed by addition of pre-diluted primary antibody and incubation for 2 h on an orbital shaker, washing with 1X wash buffer and addition of 1X blocking reagent. After 1h incubation, secondary antibody enzyme conjugate was added to each well followed by three washings and addition of substrate solution. Enzyme reaction was stopped by addition of 100 μl stop solution as soon as the color started to appear. The results were read immediately on spectrophotometer using 450 nm as primary wavelength and standard curve was prepared using standard 5-methyl-2-deoxycytidine concentrations versus corresponding absorbance values. The absorbance values of unknown samples were subsequently used to calculate the concentration of 5-Methyl-2-deoxycytidine within their gDNAs. The samples were analyzed for 5-methyl-2-deoxycytidine concentrations based on mean absorbance values of three independent measurements, each taken in duplicate.
Statistical analysis
qPCR data for KoSR vs FBS based differentiation media were compared by Student’s t-test using a statistical software program (SPSS 11.5, 2004, IBM, USA). GraphPad PRISM version 4 (GraphPad Software Inc., San Diego CA) was used for building standard curve and calculation of 5-methyl-2-deoxycytidine concentrations directly from the corresponding absorption values. The results are expressed as mean ± S.E. of mean and the statistical significance was accepted at p <0.05.
Results
qPCR analysis for germ lineage genes
qPCR expression profiles are shown in Figure 1 for both KoSR and FBS generated EBs, taking undifferentiated ES cells as the calibrator. NANOG showed decrease in expression throughout the culture period (normalized expression less than 1) as compared to the calibrator (normalized expression equal to 1) under both the differentiation conditions. OCT4 showed a six-fold increased expression in day 8 EBs produced in KoSR differentiation medium while the corresponding expression in FBS based medium was lesser than the control. DAZL showed significantly (p<0.05) increased expression (2.5 fold greater than control) in day 14 EBs obtained from FBS based spontaneous differentiation medium, while the corresponding expression in day 4 and day 8 EBs revealed no significant difference between KoSR and FBS based media. VASA showed significantly increased expression (3-fold higher) in day 4 EBs obtained in KoSR based medium as compared to the corresponding EBs from FBS based medium. Its expression although greater than control, showed no significant (p<0.05) difference in day 8 and day 14 EBs, differentiated in KoSR and FBS based media. PLZF did not show any significant change in expression pattern throughout the culture interval in both types of differentiation media. SYCP3 showed significantly greater expression in KoSR-based media, throughout the culture interval, with the highest expression in day 8 EBs. MLH1 showed highest expression in day 4 EBs obtained from FBS based medium, thereafter showing a decreasing trend as the culture interval prolonged. Its expression was significantly greater in day 8 EBs obtained from KoSR based SSCs, while day 14 EBs did not show any significant (p<0.05) difference in expression profile between the two media types. PRM2 expression was highest in day 8 EBs obtained from both the media formulations in comparison to the corresponding day 4 and day 14 EBs, with the highest expression in EBs produced in FBS based medium. The expression of BOULE was significantly (p<0.0) greater in FBS based media throughout the culture interval, while TEKT1 expression showed a significant increase in day 14 EBs obtained from KoSR supplemented medium. Day 4 EBs derived from FBS based medium showed a significantly (p<0.05) greater expression of GDF9, unlike day 8 EBs which showed significantly increased expression in KoSR based media. 14 day old EBs did not show any significant (p<0.05) difference in expression pattern of GDF9 between the two media types. ZP2 expression was highest (2-3 fold) in day 8 EBs as compared to the corresponding day 4 and day 14 ones. The expression, however, was not significantly different between the two media types while ZP3 showed increased expression, though not significant, in KoSR-based EBs analyzed at day 4 and 14 of the culture (Figure 1). Hence, it could be concluded that ES cells do express various germ linage associated markers upon spontaneous differentiation.
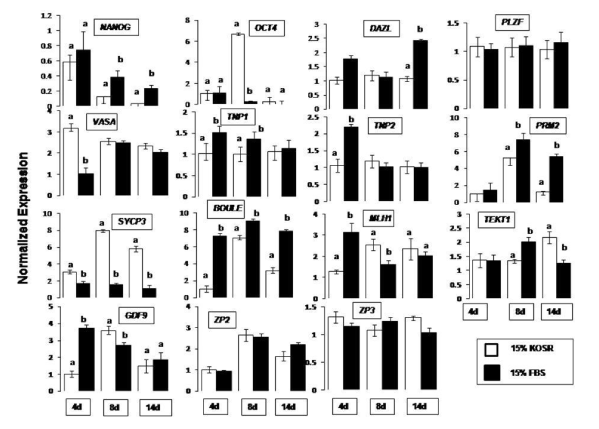
Figure 1. qPCR analysis of germ lineage marker expression in EBs collected at day 4, 8 and 14 of culture interval, upon spontaneous differentiation in KoSR- and FBS- based media. Bars represent 2-ΔΔcT values ± S.E. of mean, and are calibrated against the corresponding normalized values of undifferentiated ES Cell colonies. Bars with different superscripts differ significantly (p<0.05).
Immunocytochemical analysis for germ lineage proteins
Based on qPCR data analysis, immunocytochemical examination of various germ lineage associated proteins was performed on both day 14 EBs as well as monolayer adherent cultures. Immunocytochemistry revealed expression of: i) PGC markers like c-Kit, Dazl and Vasa (Figure 2); ii) Meiotic proteins like Sycp3, Mlh1 and Protamine1 (Figure 3); iii) Spermatocyte markers like Acrosin and Haprin (Figure 4); and iv) Oocyte specific markers like Gdf9 and Zp4 (Figure 5) in EBs collected from both KoSR and FBS-based media formulations. The number of EBs that stained positive for various germ lineage associated proteins is given in table 1. Only 3 monolayer adherent cultures revealed the positive expression of PGC-specific proteins out of the 45 cultures that were analyzed (Figure 6). None of the cultures, both from KoSR-and FBS-based spontaneous differentiation cultures, showed the presence of meiotic and mature germ cell proteins upon immunocytochemical analysis. No such protein detection was observed in undifferentiated (control) ES colonies and monolayer cultures (Figure S1 and S2)
KoSR-based spontaneous differentiation |
Germ lineage protein |
c-KIT |
DAZL |
VASA |
MLH1 |
SYCP3 |
PROTAMINE1 |
ACROSIN |
HAPRIN |
GDF9 |
ZP4 |
Total EBs analyzed |
24 |
24 |
24 |
30 |
30 |
30 |
35 |
35 |
34 |
34 |
EBs expressing the protein |
6
(25%) |
7
(29.2%) |
8
(33.3%) |
4
(13.3%) |
3
(10%) |
3
(10%) |
3
(8.6%) |
4
(11.4%) |
5
(14.7%) |
4
(11.8%) |
FBS-based spontaneous differentiation |
Germ lineage protein |
c-KIT |
DAZL |
VASA |
MLH1 |
SYCP3 |
PROTAMINE1 |
ACROSIN |
HAPRIN |
GDF9 |
ZP4 |
Total EBs analyzed |
24 |
24 |
24 |
30 |
30 |
30 |
35 |
35 |
34 |
34 |
EBs expressing the protein |
6
(25%) |
6
(25%) |
6
(25%) |
3
(10%) |
2
(6.7%) |
2
(6.7%) |
2
(5.7%) |
1
(2.8%) |
3
(8.8%) |
2
(5.9%) |
Table 1. Immunocytochemical analysis results for germ lineage gene induction in embryoid bodies derived from KoSR- and FBS-based spontaneous differentiation media.
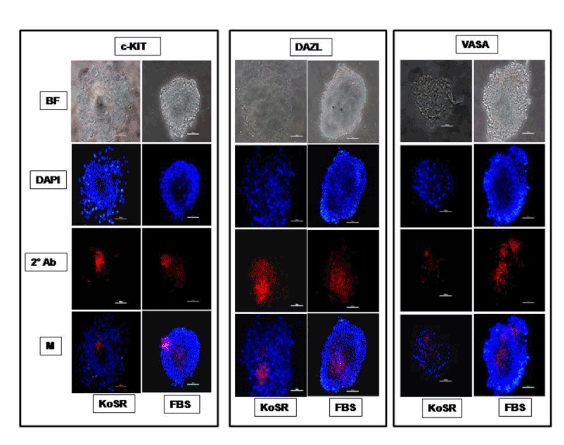
Figure 2. Immunocytochemical characterization of day 14 EBs derived via Static Suspension Culture in KoSR and FBS- based spontaneous differentiation media for Primordial germ cell marker (c-Kit, Dazl and Vasa) expression; BF - Bright field image; DAPI - Nuclear stained image; 2˚Ab - PE/ FITC labeled secondary antibody image; M - Merged images of DAPI and 2˚Ab (Magnification- 200X, Scale bar-100µm).
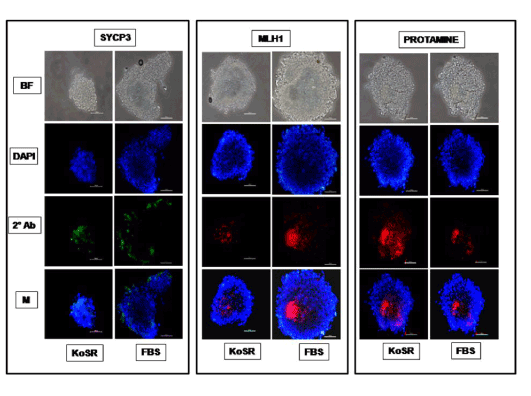
Figure 3. Immunocytochemical characterization of day 14 EBs derived from KoSR- and FBS- based spontaneous differentiation media for Meiotic marker (Sycp3, Mlh1 and Protamine1) expression; BF - Bright field image; DAPI - Nuclear stained image; 2˚Ab - PE/ FITC labeled secondary antibody image; M - Merged images of DAPI and 2˚Ab (Magnification- 200X, Scale bar-100µm).
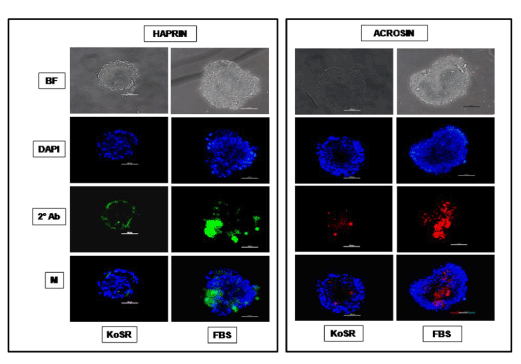
Figure 4. Immunocytochemical characterization of day 14 EBs developed in KoSR- and FBS- based spontaneous differentiation media for Spermatocyte marker (Acrosin and Haprin) expression; BF - Bright field image; DAPI - Nuclear stained image; 2˚Ab - PE/ FITC labeled secondary antibody image; M - Merged images of DAPI and 2˚Ab (Magnification- 200X, Scale bar-100µm).
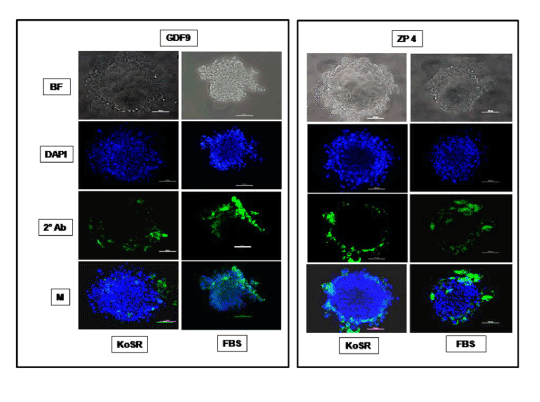
Figure 5. Immunocytochemical characterization of day 14 EBs derived from KoSR- and FBS- based spontaneous differentiation media for Oocyte marker (Gdf9 and Zp4) expression; BF - Bright field image; DAPI - Nuclear stained image; 2˚Ab - PE/ FITC labeled secondary antibody image; M - Merged images of DAPI and 2˚Ab (Magnification- 200X, Scale bar-100µm).
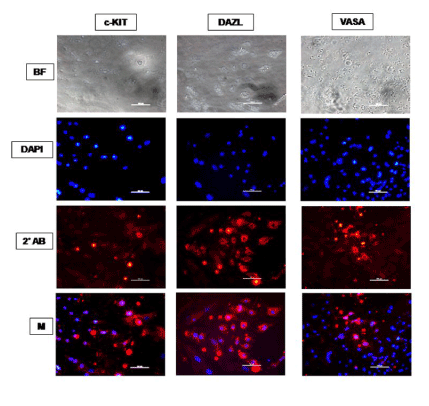
Figure 6. Immunocytochemical characterization of ES cell monolayer adherent cultures differentiated spontaneously for 14 days in KoSR-based media for Primordial germ cell marker (c-Kit, Dazl and Vasa) expression; BF - Bright field image; DAPI - Nuclear stained image; 2˚Ab - PE/ FITC labeled secondary antibody image; M - Merged images of DAPI and 2˚Ab (Magnification- 200X, Scale bar-100µm).
Global DNA methylation analysis
Global DNA methylation analysis did not reveal any significantly (p<0.05) different 5-methyl-2-deoxycytidine concentrations in day 14 EBs from the control ES cells (Figure 7), indicating no significant initiation of the methylation erasure process.
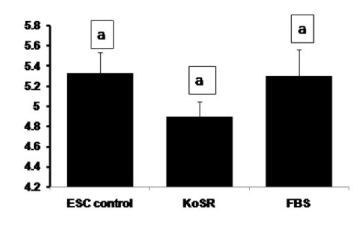
Figure 7. Global DNA methylation analysis for 5-Methyl-2-deoxycytidine concentration of undifferentiated ES cell colonies and EBs differentiated for 14 days in KoSR- and FBS- based spontaneous differentiation media. Bars with same superscripts do not differ significantly (p<0.05).
Discussion
Assaying ES cell differentiation to germ lineage
A fairly good number of reports exist which demonstrate that ES cells, irrespective of the species of origin, are pluripotent and capable of differentiating into a wide variety of cell types under appropriate culture conditions [9,20]. However, very scanty information is available on differentiation of embryonic stem cells into germ cells, apart from some reports in mice and human embryonic and/or induced pluripotent stem cells [8,9,11,21,22]. However, germ lineage differentiation from bubaline ES cells or stem cell-like cells has not been reported and, as per our knowledge, this is the first draft reporting spontaneous differentiation of bubaline embryonic stem cells into germ lineage cells. For this purpose, we studied a panel of germ cell-specific/ enriched markers together with embryonic stem cell markers to model in vitro differentiation of ES cells into germ lineage cells. There are a number of evidences which support the validity of this approach to assay germ cell development [9,23,24]. A belief, though untrue, was held that ES cells express all the genes promiscuously thus annulling cell-specific marker analysis as inappropriate means to monitor their differentiation into any given cell type. However, a number of reports have demonstrated that ES cells do not express all genes in a promiscuous manner [25-29], thereby validating the marker-based identification approach. We also observed a non-random pattern of gene expression in our study which began with expression of early markers of germ line-specific/ enriched genes (PGC-specific markers) at lesser culture intervals, while markers associated with meiosis and spermatocyte as well as oocyte development appeared to show appreciable expression at later stages of culture interval. We also connected transcription of germ cell-specific genes with translation. The reports that only germ line cells express proteins like Dazl, Vasa, Sycp3, Gdf9, Tekt1, Tnps, Prms, Acrosin, Boule, Zp4, Haprin etc. [9,11,14,22] further support our model to investigate ES cell differentiation into germ lineage cells.
Transcriptional profiling of germ lineage genes
It has been reported that ES cells are reprogrammed spontaneously to go through germ lineage, in addition to their differentiation into many other lineages. Such reprogramming has been reported in mice when murine ES cells were cultured in absence of pluripotency maintaining factors like rmLIF and feeder cells [23,24,30,31]. Spontaneous differentiation to germ line cells has been reported in murine as well as human ES cells in adherent cultures after removal of pluripotency factors like rmLIF and bFGF [32,33] as well as through formation of embryoid bodies [30,31,34]. Upon spontaneous differentiation through EB culture, we observed significantly (p<0.05) increased expression of germ lineage associated genes in EBs collected at 4, 8 and 14 days of culture interval as compared to the control ES cell colonies. The highest expression of the genes was more or less observed in 8 day old EBs in both types of differentiation cultures. FBS based spontaneous differentiation cultures induced greater expression of genes like DAZL, TNP1, PRM2 and BOULE as compared to KoSR-based spontaneous cultures, probably, due to presence of a large number of growth factors present in FBS as compared to KoSR, which is a commercial serum replacer, designed primarily for maintaining stem cells in an undifferentiated state under normal culture conditions. The active ingredient in serum replacer is a lipid- rich albumin which, unlike albumin of FBS, has a strong positive effect on the expansion of undifferentiated human ES cells in culture [35]. The chemical nature of FBS owing to its defined and/or undefined components could also lead to random and non-tailored differentiation of the pluripotent cells across different lineages.
Immuocytochemical analysis for germ lineage markers
Based on the germ lineage gene expression data from qPCR, we collected day 14 EBs for immunocytochemistry. These EBs expressed PGC-specific markers (c-Kit, Dazl and Vasa); Meiotic markers (Sycp3, Mlh1 and Protamine); Spermatocyte-specific markers (Acrosin and Haprin); as well as Oocyte-specific markers (Gdf9 and Zp4); indicating differentiation into germ lineage-like cells. However, MaxGel-adherent differentiated monolayer showed only expression of PGC-specific markers like c-Kit, Dazl and Vasa, indicating that the differentiation has not progressed into meiotic and later germ lineage events. Since, germ cell differentiation process is dependent on spatial distribution of differentiating cells, the three-dimensional structure as represented by EBs probably provide a microenvironment that enhances germ lineage differentiation, although other cell types besides germ cells may also arise from them [36]. As per this observation, EB culture provides a better alternative than adherent culture for inducing germ lineage differentiation but the most pertinent problem with EB culture is sorting of the differentiated cells from EBs, since they grow as tight three-dimensional aggregates, unlike monolayer cultures which could be readily disaggregated by enzymatic treatment.
Global DNA methylation analysis
It is a well-known fact that epigenetic modifications are sequentially established and erased in germ lineage. PGCs undergo DNA demethylation of the imprinted loci to erase the parental imprints [37,38]. The imprinting process is mainly achieved by demethylation of the cytosine residues in CpG islands. It has been reported that ES cells retain the ability to carry out global de novo methylation unlike their somatic counterparts, but at the same time are capable of distinguishing and protecting CpG islands from this process [39], thereby ensuring their self-renewal and unspecialized (undifferentiated) nature. Though 5-methyl-2-deoxycytidine concentration was not significantly different in undifferentiated ES cells and day 14 EBs collected from KoSR- and FBS-based spontaneous differentiation cultures, the concentration was nevertheless lower in EBs collected from KoSR based spontaneous cultures, indicating comparatively greater methylation erasure.
To our knowledge, this is the first study of its type in farm animals aiming at differentiation of bubaline ES cells into germ line cells under spontaneous differentiation milieu. We observed that KoSR- based spontaneous medium would be better in inducing differentiation to germ lineage than FBS- based medium which contains a host of undefined chemical components that might hinder progression into meiosis and post-meiotic germ cell development. The addition of exogenous factors that are capable of inducing germ lineage differentiation should, hence, be attempted in KoSR-based medium and not in FBS-based medium. EB culture also revealed to be more efficient in inducing differentiation than monolayer culture but the problem lies in isolation and sorting of the differentiated cells from these three-dimensional aggregates. The obvious solution would be to attempt 3-dimensional culture strategies for ES cell monolayer culture which would combine both the advantages of EB culture and the ease of sorting for differentiated cells. It could be concluded that ES cells can be differentiated into germ lineage cells under in vitro culture and there is a sound possibility of generating the mature and functionally competent germ cells in vitro. Such studies will also enhance our knowledge of farm animal gametogenesis especially of bubaline species. The future implications would be to develop the strategies, whether nutritional, managerial, chemical or biotechnological, which would enhance the quality and quantity of the gametes, reduce the age of sexual maturity which currently goes upto 2-3 years in buffalo, and develop and design the gametes of choice for transgenic buffalo production and conservation of endangered species by development of artificial gametes.
Conflict of interest
The authors declare no2021 Copyright OAT. All rights reserv
References
- Strome S, Lehmann R (2007) Germ versus soma decisions: lessons from flies and worms. Science 316: 392-393. [Crossref]
- Jin Z, Xie T (2006) Germline specification: small things have a big role. Curr Biol 16: R966-967. [Crossref]
- Baughman JM, Geijsen N (2005) In vitro generation of germ cells: new techniques to solve current issues. Ann N Y Acad Sci 1061: 33-40. [Crossref]
- Saitou M, Barton SC, Surani MA (2002) A molecular programme for the specification of germ cell fate in mice. Nature 418: 293-300. [Crossref]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, et al. (2006) Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol 8: 623-630.[Crossref]
- Bortvin A, Goodheart M, Liao M, Page DC (2004) Dppa3 / Pgc7 / stella is a maternal factor and is not required for germ cell specification in mice. BMC Dev Biol 4: 2. [Crossref]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, et al. (2007) PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9: 64-71. [Crossref]
- Geijsen N, Horoschak M, Kim K, Gribnau J, Eggan K, et al. (2004) Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 427: 148-154. [Crossref]
- Clark AT, Bodnar MS, Fox M, Rodriquez RT, Abeyta MJ, et al. (2004) Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet 13: 727-739. [Crossref]
- Marques-Mari AI, Lacham-Kaplan O, Medrano JV, Pellicer A, Simón C (2009) Differentiation of germ cells and gametes from stem cells. Hum Reprod Update 15: 379-390. [Crossref]
- Aflatoonian B, Ruban L, Jones M, Aflatoonian R, Fazeli A et al. (2009) In vitro post-meiotic germ cell development from human embryonic stem cells. Hum Reprod 24: 3150-3159. [Crossref]
- West FD, Machacek DW, Boyd NL, Pandiyan K, Robbins KR, et al. (2008) Enrichment and differentiation of human germ-like cells mediated by feeder cells and basic fibroblast growth factor signaling. Stem Cells 26: 2768-2776. [Crossref]
- Tilgner K, Atkinson SP, Golebiewska A, Stojkovic M, Lako M, et al. (2008) Isolation of primordial germ cells from differentiating human embryonic stem cells. Stem Cells 26: 3075-3085. [Crossref]
- Shah SM, Saini N, Ashraf S, Zandi M, Singh MK, et al. (2015) Comparative Expression Analysis of Gametogenesis-Associated Genes in Foetal and Adult Bubaline (Bubalus bubalis) Ovaries and Testes. Reprod Domest Anim 50: 365-377. [Crossref]
- Wei W, Qing T, Ye X, Liu H, Zhang D, et al. (2008) Primordial germ cell specification from embryonic stem cells. PLoS One 3: e4013. [Crossref]
- Shah SM, Saini N, Ashraf S, Zandi M, Manik RS, et al. (2015) Development, Characterization, and Pluripotency Analysis of Buffalo (Bubalus bubalis) Embryonic Stem Cell Lines Derived from In Vitro-Fertilized, Hand-Guided Cloned, and Parthenogenetic Embryos. Cell Reprogram 17: 306-322. [Crossref]
- Shah SM, Saini N, Singh MK, Manik R, Singla SK, et al. (2016) Testicular cell-conditioned medium supports embryonic stem cell differentiation toward germ lineage and to spermatocyte- and oocyte-like cells. Theriogenology. [Crossref]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101-1108. [Crossref]
- Shah SM, Saini N, Ashraf S, Singh MK, Manik RS, et al. (2015) Cumulus cell-conditioned medium supports embryonic stem cell differentiation to germ cell-like cells. Reprod Fertil Dev. [Crossref]
- Odorico JS, Kaufman DS, Thomson JA (2001) Multilineage differentiation from human embryonic stem cell lines. Stem Cells 19: 193-204. [Crossref]
- Okita K, Ichisaka T, Yamanaka S (2007) Generation of germline-competent induced pluripotent stem cells. Nature 448: 313-317. [Crossref]
- Kee K, Angeles VT, Flores M, Nguyen HN, Reijo Pera RA (2009) Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature 462: 222-225. [Crossref]
- Hübner K, Fuhrmann G, Christenson LK, Kehler J, Reinbold R, et al. (2003) Derivation of oocytes from mouse embryonic stem cells. Science 300: 1251-1256. [Crossref]
- Toyooka Y, Tsunekawa N, Akura R, Noce T (2003) Embryonic stem cells can form germ cells in vitro. Proc Natl Acad Sci U S A 100: 11457-11462. [Crossref]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, et al. (2002) A stem cell molecular signature. Science 298: 601-604. [Crossref]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA (2002) "Stemness": transcriptional profiling of embryonic and adult stem cells. Science 298: 597-600. [Crossref]
- Tanaka TS, Kunath T, Kimber WL, Jaradat SA, Stagg CA, et al. (2002) Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res 12: 1921-1928. [Crossref]
- Hsiao LL, Dangond F, Yoshida T, Hong R, Jensen RV, et al. (2001) A compendium of gene expression in normal human tissues. Physiol Genomics 7: 97-104. [Crossref]
- Warrington JA, Nair A, Mahadevappa M, Tsyganskaya M (2000) Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol Genomics 2: 143-147. [Crossref]
- Chen HF, Kuo HC, Chien CL, Shun CT, Yao YL, et al. (2007) Derivation, characterization and differentiation of human embryonic stem cells: comparing serum-containing versus serum-free media and evidence of germ cell differentiation. Hum Reprod 22: 567-577. [Crossref]
- Qing T, Shi Y, Qin H, Ye X, Wei W, et al. (2007) Induction of oocyte-like cells from mouse embryonic stem cells by co-culture with ovarian granulosa cells. Differentiation 75: 902-911. [Crossref]
- Nayernia K, Lee JH, Drusenheimer N, Nolte J, Wulf G, et al. (2006) Derivation of male germ cells from bone marrow stem cells. Lab Invest 86: 654-663. [Crossref]
- Novak I, Lightfoot DA, Wang H, Eriksson A, Mahdy E, et al. (2006) Mouse embryonic stem cells form follicle-like ovarian structures but do not progress through meiosis. Stem Cells 24: 1931–1936. [Crossref]
- Lacham-Kaplan O, Chy H, Trounson A (2006) Testicular cell conditioned medium supports differentiation of embryonic stem cells into ovarian structures containing oocytes. Stem Cells 24: 266-273. [Crossref]
- Garcia-Gonzalo FR, Izpisúa Belmonte JC (2008) Albumin-associated lipids regulate human embryonic stem cell self-renewal. PLoS One 3: e1384. [Crossref]
- Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, et al. (2000) Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med 6: 88-95. [Crossref]
- Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, et al. (2002) Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 129: 1807-1817. [Crossref]
- Yamazaki Y, Mann MR, Lee SS, Marh J, McCarrey JR, et al. (2003) Reprogramming of primordial germ cells begins before migration into the genital ridge, making these cells inadequate donors for reproductive cloning. Proc Natl Acad Sci U S A 100: 12207-12212. [Crossref]
- Ludwig G, Nejman D, Hecht M, Orlanski S, Abu-Remaileh M, et al. (2014) Aberrant DNA methylation in ES cells. PLoS One 9: e96090. [Crossref]