Abstract
In the context of an alarming frequency of gestational diabetes mellitus (GDM) observed recently in Bangladesh and a reported stronger association of genetic variants of Transcription factor 7-like 2 (TCF7L2) rs7903146 with risk of GDM, the frequency of this single nucleotide polymorphism (SNP) was studied in Bangladeshi woman. Pregnant women with no past history of glucose intolerance (N=100; age 26.22 ± 4.56 years; body mass index (BMI) 26.39 ± 3.85; mean ± SD; GDM=50, normal glucose tolerance (NGT)=50 according to WHO 2013 criteria) were recruited for the study. TCF7L2 rs7903146 polymorphism was genotyped using Sanger sequencing technique. Mothers with GDM had significantly higher age (27.54 ± 4.45 vs. 24.90 ± 4.33 years, p=0.003; mean ± SD) and BMI (27.15 ± 3.81 vs. 25.62 ± 3.77 kg/m2, p=0.047; mean ± SD) than those of mothers with NGT. Frequency of family history of DM in 1st degree relatives was also significantly higher in women with GDM (50% vs. 22%, p=0.006). CC, CT and TT genotype frequencies of the TCF7L2 rs7903146 varied between women with GDM and NGT (GDM vs. NGT, CC-CT-TT: 56, 40 and 4% vs. 70, 26 and 4% respectively; p=0.323). The T-allele frequency was higher in GDM whereas C-allele frequency in NGT (GDM vs. NGT, T-allele: 24% vs. 17%; C-allele 76% vs. 83%; p=0.220). Women with CT-genotype had a 1.9-fold (95% CI 0.82-4.53, p=0.133) and TT-genotype had a 1.2-fold (95% CI 0.17-9.44, p=0.828) increased risk for GDM. It is concluded that the TCF7L2 rs7903146 variant might confer an increased risk of GDM in Bangladeshi women.
Key words
TCF7L2 gene, GDM, Bangladesh
Introduction
Gestational diabetes mellitus (GDM) is an emerging issue for health care professionals. It is associated with adverse pregnancy outcome. In Bangladesh, an alarming frequency of GDM has been observed recently [1,2]. Despite multiple environmental risk factors, susceptibility to GDM has a strong genetic component [3-7]. Genetic predisposition may vary among the different ethnic origins [6-10]. Therefore, genetic association studies need be replicated in people of different ethnicities. Among various genetic polymorphisms, association of transcription factor 7 like 2 (TCF7L2) polymorphism with GDM has been observed in various ethnicities [5-7,11-13]. In a systematic review, genetic variant of TCF7L2 was observed to have the strongest association with GDM risk [14].
TCF7L2 seems to be required to maintain glucose stimulated insulin secretion and β-cell survival as is observed by the fact that its deletion in human islets reduces insulin secretion and increases apoptosis [15]. In the absence of TCF7L2, Ca2+ increases in the wrong part of the β-cell resulting in impairment of insulin secretion [16]. Mitchell et al. also observed that this particular gene product is required for the normal function of β-cells and their expansion under a situation of metabolic stress [17]. The most associated single nucleotide polymorphism (SNP), rs7903146 is found not in coding sequence but in an intronic region. A potential mechanism through which it may confer an increased risk of diabetes is by altering TCF7L2 expression levels [18].
In this perspective, the present study is aimed to investigate association of TCF7L2 gene polymorphism rs7903146 with GDM in Bangladeshi pregnant women.
Materials and methods
Study subjects
This cross-sectional study included Bangladeshi pregnant women with no past history of glucose intolerance [N=100; age 26.22 ± 4.56 years; body mass index (BMI) 26.39 ± 3.85 kg/m2; mean ± SD] from the antenatal clinics of Bangabandhu Sheikh Mujib Medical University (BSMMU), Dhaka, Bangladesh during the period of January to December, 2014. They were screened by a 03-sample 75-gm oral glucose tolerance test (OGTT) following World Health Organization (WHO) 2013 criteria regardless of weeks of gestation [19]. Mothers with normal OGTT, if performed before 24 weeks of gestation, underwent repeat test between 24-28th weeks of pregnancy to assign the glycemic status. Any subject falling into the group of ‘diabetes in pregnancy’ according to WHO 2013 criteria was excluded. TCF7L2 rs7903146 polymorphism was genotyped in 50 GDM patients and equal number of normal glucose tolerant (NGT) mothers. Relevant clinical and biochemical data including hemoglobin A1c (HbA1c) and plasma glucose (PG) were recorded (Table 1).
Table 1. Characteristics of the study subjects. Significance values stand for comparison between GDM and NGT by Student’s t-test and χ2-test
Variables |
All subjects |
GDM |
NGT |
p |
N |
100 |
50 |
50 |
|
Age in years (mean ± SD) |
26.22 ± 4.56 |
27.54 ± 4.45 |
24.90 ± 4.33 |
0.003 |
BMI in kg/m2 (mean ± SD) |
26.39 ± 3.85 |
27.15 ± 3.81 |
25.62 ± 3.77 |
0.047 |
Occupation |
Housewife |
66 (66%) |
34 (68%) |
32 (64%) |
0.839 |
Service |
11 (11%) |
06 (12%) |
05 (10%) |
Medical professional |
13 (13%) |
05 (10%) |
08 (16%) |
Student |
10 (10%) |
05 (10%) |
05 (10%) |
Parity |
Primipara |
41 (41%) |
17 (34%) |
24 (48%) |
0.222 |
Multipara |
59 (59%) |
33 (66%) |
26 (52%) |
History of abortion |
29 (29%) |
14 (28%) |
15 (30%) |
1.000 |
SBP in mm Hg (mean ± SD) |
101.35 ± 10.49 |
102.30 ± 11.66 |
100.40 ± 9.19 |
0.368 |
DBP in mm Hg (mean ± SD) |
63.15 ± 9.01 |
64.30 ± 10.00 |
62.00 ± 7.82 |
0.203 |
Family history of DM in 1st degree relatives |
36 (36%) |
25 (50%) |
11 (22%) |
0.006 |
GDM: Gestational Diabetes Mellitus; NGT: Normal Glucose Tolerance; BMI: Body Mass Index; DM: Diabetes Mellitus; SBP: Systolic Blood Pressure; DBP: Diastolic Blood Pressure
Written informed consent was taken from the participants. The project was run after approval of the institutional review board (IRB) of BSMMU.
Assay methods
Plasma glucose was assayed by glucose-oxidase method while HbA1c was measured using the Bio-Rad D-10TM HbA1c Program 220-0101 (USA) certified by National Glycohemoglobin Standardization Program. Inter-assay co-efficient variance (CV) for glucose was 4.44%.
Genotyping
Approximately 3.0 ml peripheral blood samples in duplicate were collected in a VACUETTE® EDTA K3 (Greiner Bio-One GmbH) tube, from the participants. Genomic DNA was extracted using QIAmp DNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany). Extracted DNA was quantified by Quantusâ fluorometer (Promega Corporation, USA). TCF7L2 locus was amplified by polymerase chain reaction (PCR) using commercial PCR kit (GoTaq, Promega Corporation, USA). The PCR primers were designed by Primer Express and optimized according to the manufacturer's protocol. Forward and reverse primers used were 5/AGATTCCTTTTTAAATGGTGACA3/ and 5/GCATTACAAATTATTAGAACTTTCA3/. The thermocycling conditions consisted of one denaturing cycle at 95°C for 10 minutes followed by 35 cycles of denaturing at 95°C for 30 seconds, annealing at 52.0°C for 1 minute, and extension at 72°C for 1 minute. Final extension was at 72°C for 10 minutes. The amplicons were then electrophoresed in a 2% agarose gel to assess PCR efficacy and to detect the presence of the 356-bp of TCF7L2 locus (Figure 1).
Figure 1. Detection of amplified region of TCF7L2 locus (356 bp) in 2% agarose gel electrophoresis.
M- 100 bp DNA ladder marker;
Lane 2 – 9 : Amplicons from 8 different participants.
Sanger’s di-deoxy chain terminating method was used to sequence the amplified TCF7L2 locus. Chain terminating cycle sequencing for both forward and reverse primer was performed using sequencing kit (BigDye Terminator v3.1, Applied Biosystems, Foster City, Calif. USA). Sequencing primers were same as amplification ones. Capillary electrophoresis was performed using ABI 3500Dx Genetic Analyzer (Applied Biosystems, Foster City, Calif. USA). Sequences were analyzed to determine the genotype using SeqScape software (Applied Biosystems, Foster City, Calif. USA) (Figure 2). Genotyping results were validated by re-sequencing 10% of randomly selected samples. No differences were found, thus the genotyping error rate was 0%.
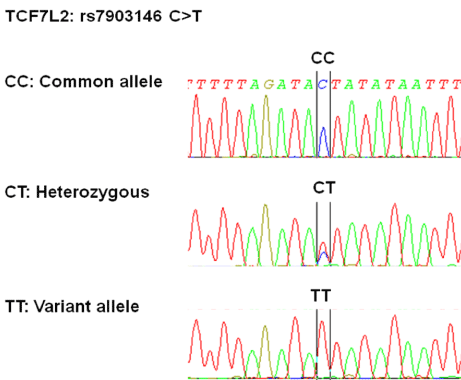
Figure 2. Representative sequencing of PCR products amplified with primers of rs7903146. CC: common allele; CT: variant heterozygous; TT: variant homozygous allele
Statistics
Data of genotypes (CC, CT, TT) were expressed as frequencies and/or percentages. Mean (± SD) values of parametric data were compared by unpaired Student’s t-test. Hardy-Weinberg equilibrium (HWE) test was performed to see the distribution of variant allele genotype among normal population. Association of genotypes with glycemic groups was assessed by χ2-test and/or odds ratio (OR). P values £ 0.05 were considered as significant.
Results
Clinical and glycemic profile of the study subjects
Mothers with GDM had significantly higher age (27.54 ± 4.45 vs. 24.90 ± 4.33 years, p=0.003; mean ± SD) and BMI (27.15 ± 3.81 vs. 25.62 ± 3.77 kg/m2, p=0.047; mean ± SD) than those of mothers with NGT. Frequency of family history of DM in 1st degree relatives was also significantly higher in women with GDM (50% vs. 22%, p=0.006). However, none of occupation (p=0.839), parity (p=0.222), history of abortion (p=1.000), systolic (p=0.368) or diastolic (p=0.203) blood pressure were statistically different between the two groups (Table 1).
TCF7L2 rs7903146 polymorphism in GDM & non-GDM woman
Genotyping for TCF7L2 rs7903146 polymorphism revealed that CC, CT and TT genotype distribution of the TCF7L2 rs7903146 differed between women with GDM and NGT (GDM vs. NGT, CC-CT-TT: 56, 40 and 4% vs. 70, 26 and 4% respectively, p=0.323). The T-allele frequency was higher in GDM whereas C-allele frequency in NGT (GDM vs. NGT, T-allele: 24% vs. 17%; C-allele 76% vs. 83%; p=0.220) (Table 2). This genotype distribution was consistent with Hardy-Weinberg equilibrium (χ2=0.309; p = 0.573). Mothers having CT-genotype had higher frequency of GDM (CT vs. CC: 60.6% vs. 44.4%, p=0.133) with a 1.9-fold (95% CI 0.82-4.53) increased risk of having GDM, whereas TT-genotype had a 1.2-fold (95% CI 0.17-9.44) increased risk as compared with CC genotype (TT vs. CC: 50.0% vs. 44.4%, p=0.828). Comparison in dominant model (CC vs. CT/TT: 44.4% vs. 59.5%, p=0.147) demonstrated an OR of 1.8 (95% CI 0.81-4.18). But when compared in recessive model (CT/CC vs. TT), no increased risk was observed (OR 1.00, 95% CI: 0.135-7.392) (Table 3).
Table 2. Variants of TCF7L2 rs7903146 in study subjects
Variables |
GDM |
NGT |
Total |
χ2, p |
Genotype |
CC |
28 (56%) |
35 (70%) |
63 (63%) |
χ2=2.263
p=0.323 |
CT |
20 (40%) |
13 (26%) |
33 (33%) |
TT |
02 (04%) |
02 (04%) |
04 (04%) |
Total |
50 |
50 |
100 |
|
Allele frequency |
Minor allele frequency (T) |
24 (24%) |
17 (17%) |
41 (20.5%) |
χ2=1.503
p=0.220 |
Major allele frequency (C) |
76 (76%) |
83 (83%) |
159 (79.5%) |
Total |
100 |
100 |
200 |
|
TCF7L2: Tranascription Factor 7 Like 2; GDM: Gestational Diabetes Mellitus; NGT: Normal Glucose Tolerance; C: Cytosine; T: Thymine
Table 3. Comparison of glycemic outcome among the variants of TCF7L2 rs7903146 in study subjects
Compared groups |
N |
GDM |
NGT |
OR for GDM (95% CI) |
p |
CC vs. CT |
CC |
63 |
28 (44.4%) |
35 (55.6%) |
1.923
(0.816-4.531) |
0.133 |
CT |
33 |
20 (60.6%) |
13 (39.4%) |
CC vs. TT |
CC |
63 |
28 (44.4%) |
35 (55.6%) |
1.250
(0.165-9.442) |
0.828 |
TT |
4 |
2 (50.0%) |
2 (50.0%) |
CT/CC vs. TT |
CT/CC |
96 |
48 (50.0%) |
48 (50.0%) |
1.000
(0.135-7.392) |
1.000 |
TT |
4 |
2 (50.0%) |
2 (50.0%) |
CC vs. CT/TT |
CC |
63 |
28 (44.4%) |
35 (55.6%) |
1.8333
(0.805-4.176) |
0.147 |
2021 Copyright OAT. All rights reserv
CT/TT |
37 |
22 (59.5%) |
15 (40.5%) |
TCF7L2: Tranascription Factor 7 Like 2; GDM: Gestational Diabetes Mellitus; NGT: Normal Glucose Tolerance; C: Cytosine; T: Thymine
Discussion
In the present study, SNP of TCF7L2 rs7903146 was studied in Bangladeshi GDM mothers. It was observed that the CC, CT and TT genotype frequencies varied between women with GDM and NGT with a 1.9-fold increased risk of GDM in women with CT in comparison to those with CC genotype. Homozygous TT variants are expected to have higher risk for GDM than that of heterozygous CT as demonstrated in other studies5. However, present study failed to distinguish higher risk for subjects with TT variant who were very few in number (n=4, GDM=2, NGT=2). However, T-allele frequency was observed to be higher in GDM, though not reaching level of significance. Previous studies also observed that women with risk allele (T) of TCF7L2 rs7903146 had higher risk of GDM [5,11]. In fact, this genetic variant is the most widely studied variant in association with GDM with a consistent and strong association across different populations [14].
This study did not investigate the polymorphism in other loci precipitating GDM [20-22], and thus it cannot be inferred that findings of glycemic status in the studied mothers are solely attributable to the variation of investigated SNP. The 75g 3-sample OGTT, according to WHO 2013 criteria labels women with higher glycemic values as ‘DM in pregnancy’ 19. They were not included in the study as there might be pre-existing glucose intolerance in this particular group and hence may not be true GDM. We could not exclude the possibility of pre-pregnancy glucose intolerance in every case; but the lesser magnitude of hyperglycemia of ‘GDM’ group as in WHO 2013 criteria reduces such possibility. To identify the non-GDM women, subjects having done OGTT after 24 weeks of gestation were preferred and if done before 24 weeks of gestation, OGTT was repeated at or after 24 weeks in those instances. Higher age, BMI and family history of DM in 1st degree relatives are considered to be important risk factors of GDM [1,2,23]. We also observed significantly higher age, BMI and frequency of family history of DM in women with GDM. The SNP under evaluation was observed to maintain Hardy-Weinberg equilibrium.
Almost all genetic loci associated with GDM risk, including TCF7L2, have been previously related to the risk of T2DM [24-29]. The effect size of the associations between these SNPs and GDM was similar to those in the studies of T2DM [30]. These findings suggest an at least partly shared genetic basis between GDM and T2DM, which is not surprising given that both insulin resistance and defects in insulin secretion play key roles in the etiology of both GDM and T2DM. Evaluation of a single SNP in small number of subjects itself is unlikely to lead to any precise decision. But multiple studies on large-scale for identification of GDM susceptibility variants can lead to novel biological insights and improved measures of individual etiological processes [31]. Novel biological insights may lead to development of new therapeutic targets, biomarkers and opportunities for disease prevention.
In summary, the TCF7L2 rs7903146 variant is observed to be associated with an increased risk of GDM in Bangladeshi women.
Acknowledgements
The authors gratefully acknowledge the scientific colleagues of the department for their generous and moral support to the project. Supporting staffs of the department and patients participating in the project are also thanked and appreciated. Novo Nordisk Pharma Bangladesh is acknowledged for their financial help.
Disclosure
The authors declare they have no competing interests that might be perceived to influence the results and discussion reported in this paper.
References
- Sultana N, Hasanat MA, Alam K, Jahan S, Hasan M, et al. (2016) Alarming frequency of gestational diabetes mellitus (GDM) attending a tertiary care hospital in Bangadesh. J Clin Diabetol 2: 1-5.
- Panthi S, Hasanat MA, Hasan M, Aktar Y, Sultana N, et al. (2015) Frequency of gestational diabetes mellitus in Bangladesh, impact of WHO 2013 screening criteria: efficiency of DIPSI and WHO 1999 criteria. J Clin Diabetol 2: 13-19.
- Watanabe RM, Allayee H, Xiang AH, Trigo E, Hartiala J, et al. (2007) Transcription factor 7-like 2 (TCF7L2) is associated with gestational diabetes mellitus and interacts with adiposity to alter insulin secretion in Mexican Americans. Diabetes 56: 1481-1485. [Crossref]
- Cauchi S, El Achhab Y, Choquet H, Dina C, Krempler F, et al. (2007) TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med (Berl) 85: 777-782. [Crossref]
- Shaat N, Lernmark A, Karlsson E, Ivarsson S, Parikh H, et al. (2007) A variant in the transcription factor 7-like 2 (TCF7L2) gene is associated with an increased risk of gestational diabetes mellitus. Diabetologia 50: 972-979. [Crossref]
- Watanabe RM, Black MH, Xiang AH, Allayee H, Lawrence JM, et al. (2007) Genetics of gestational diabetes mellitus and type 2 diabetes. Diabetes Care 30 Suppl 2: S134-140. [Crossref]
- Kwak SH, Jang HC, Park KS (2012) Finding genetic risk factors of gestational diabetes. Genomics Inform 10: 239-243. [Crossref]
- Dornhorst A, Paterson CM, Nicholls JS, Wadsworth J, Chiu DC, et al. (1992) High prevalence of gestational diabetes in women from ethnic minority groups. Diabet Med 9: 820-825. [Crossref]
- Wang Y, Nie M, Li W, Ping F, Hu Y, et al. (2011) Association of six single nucleotide polymorphisms with gestational diabetes mellitus in a Chinese population. PLoS One 6: e26953. [Crossref]
- Vejrazkova D, Lukasova P1, Vankova M1, Vcelak J1, Bradnova O1, et al. (2014) MTNR1B Genetic Variability Is Associated with Gestational Diabetes in Czech Women. Int J Endocrinol 2014: 508923. [Crossref]
- Papadopoulou A, Lynch KF, Shaat N, Håkansson R, Ivarsson SA, et al. (2011) Gestational diabetes mellitus is associated with TCF7L2 gene polymorphisms independent of HLA-DQB1*0602 genotypes and islet cell autoantibodies. Diabet Med 28: 1018-1027. [Crossref]
- Huopio H, Cederberg H, Vangipurapu J, Hakkarainen H, Pääkkönen M, et al. (2013) Association of risk variants for type 2 diabetes and hyperglycemia with gestational diabetes. Eur J Endocrinol 169: 291-297. [Crossref]
- Kanthimathi S, Chidambaram M, Liju S, Bhavadharini B, Bodhini D, et al. (2015) Identification of genetic variants of gestational diabetes in South Indians. Diabetes Technol Ther 17: 462-467. [Crossref]
- Zhang C, Bao W, Rong Y, Yang H, Bowers K, et al. (2013) Genetic variants and the risk of gestational diabetes mellitus: a systematic review. Hum Reprod Update 19: 376-390. [Crossref]
- Shu L, Sauter NS, Schulthess FT, Matveyenko AV, Oberholzer J, et al. (2008) Transcription factor 7-like 2 regulates beta-cell survival and function in human pancreatic islets. Diabetes 57: 645-653. [Crossref]
- Gloyn AL, Braun M, Rorsman P (2009) Type 2 diabetes susceptibility gene TCF7L2 and its role in beta-cell function. Diabetes 58: 800-802. [Crossref]
- Mitchell RK, Mondragon A, Chen L, Mcginty JA, French PM, et al. (2015) Selective disruption of Tcf7l2 in the pancreatic β cell impairs secretory function and lowers β cell mass. Hum Mol Genet 24: 1390-1399. [Crossref]
- Groop L (2010) Open chromatin and diabetes risk. Nat Genet 42: 190-192. [Crossref]
- World Health Organization (2013) Diagnostic criteria and classification of hyperglycaemia first detected in pregnancy. Geneva, pp 1-63.
- Helgason A, Pálsson S, Thorleifsson G, Grant SF, Emilsson V, et al. (2007) Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 39: 218-225. [Crossref]
- Kwak SH, Kim SH, Cho YM, Go MJ, Cho YS, et al. (2012) A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes 61: 531-541. [Crossref]
- Mao H, Li Q, Gao S (2012) Meta-analysis of the relationship between common type 2 diabetes risk gene variants with gestational diabetes mellitus. PLoS One 7: e45882. [Crossref]
- Seshiah V, Balaji V, Balaji MS, Sekar A, Sanjeevi CB, et al. (2005) One step procedure for screening and diagnosis of gestational diabetes mellitus. J Obstet Gynecol India 55: 525-529.
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, et al. (2006) Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38: 320-323. [Crossref]
- Bodhini D, Radha V, Dhar M, Narayani N, Mohan V (2007) The rs12255372(G/T) and rs7903146(C/T) polymorphisms of the TCF7L2 gene are associated with type 2 diabetes mellitus in Asian Indians. Metabolism 56: 1174-1178. [Crossref]
- Amoli MM, Amiri P, Tavakkoly-Bazzaz J, Charmchi E, Hafeziyeh J, et al. (2010) Replication of TCF7L2 rs7903146 association with type 2 diabetes in an Iranian population. Genet Mol Biol 33: 449-451. [Crossref]
- Chauhan G, Spurgeon CJ, Tabassum R, Bhaskar S, Kulkarni SR, et al. (2010) Impact of common variants of PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, and CDKAL1 on the risk of type 2 diabetes in 5,164 Indians. Diabetes 59: 2068-2074. [Crossref]
- Palmer ND, Hester JM, An SS, Adeyemo A, Rotimi C, et al. (2011) Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes 60: 662-668. [Crossref]
- Danquah I, Othmer T, Frank LK, Bedu-Addo G, Schulze MB, et al. (2013) The TCF7L2 rs7903146 (T) allele is associated with type 2 diabetes in urban Ghana: a hospital-based case-control study. BMC Med Genet 14: 96. [Crossref]
- Robitaille J, Grant AM (2008) The genetics of gestational diabetes mellitus: evidence for relationship with type 2 diabetes mellitus. Genet Med 10: 240-250. [Crossref]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9: 356-369. [Crossref]


