Abstract
Much of the recent gains in knowledge regarding the influence of patient genetics on medication pharmacokinetics (drug absorption, distribution, metabolism and elimination) how patients process medications) and pharmacodynamics (drug response) have been attributed to the technologic advances in genetic testing methodologies and the involvement of large clinical data sets and biobanks. Indeed, Genome Wide Association Studies (GWAS) and Phenome Wide Association Studies (PWAS) along with ever-evolving biomedical informatics techniques and the expansion of the -omics sciences (e.g., transcriptomics, metabolomics, proteomics) have brought about unprecedented advances in precision medicine. Although the simpler candidate-gene analysis technique is not considered cutting-edge, it is reliable and important to the translation of pharmacogenomic research and the advancement of precision medicine. Leveraging the knowledge of biological plausibility (i.e., genetic mutation → altered function of protein product → altered drug pharmacokinetics/dynamics) to appropriately select genes for inclusion, the candidate-gene analysis technique does not necessitate large patient cohorts nor extensive multi-gene genetic analysis arrays. It is often the ideal method for clinicians to begin evaluating whether genetic information might improve their pharmacologic treatment strategies for their patients. Having access to specific patient populations and expertise regarding their medical subspecialty, physician scientists can implement a candidate-gene analysis in small cohorts. Even with less than 100 patients, results can often be used to determine whether further investigation is warranted and to inform future studies. Herein, we present a comparison of select contemporary methodologies regarding collection, processing and genotype testing applicable to the efficient implementation of candidate-gene studies.
Introduction
The candidate-gene analysis method is ideal for initial, exploratory genetic association studies in patient cohorts of limited size involving a limited number of polymorphisms in a select number of genes. Relying on known (or biologically plausible) mechanistic links to qualify candidate genes and polymorphisms for inclusion in the analysis, statistical power is less handicapped by substantial correction for multiple hypothesis testing (i.e., increased number of genes/polymorphism included in the analysis → increased number of hypothesis tested → increased correction/penalty for multiple hypothesis testing → increased sample size required to maintain statistical power), resulting in improved likelihood for meaningful results for analysis in smaller patient cohorts [1-4].
Disadvantages of candidate gene analysis includes reliance on previously established knowledge for disease and requiring bioinformatics and mathematical algorithms to establish candidate gene status, missed potential influences from genes that lack mechanistic links to the phenotype of interest, can only cover a limited number of genes (fewer than 1000 SNPs), has a high false positive rate and are plagued by irreproducibility. Despite these disadvantages, candidate-gene analysis can be used to complement GWAS to examine and establish links between gene status and phenotypic expression [1,4].
While studies have shown that the genotyping efficiency for saliva (total DNA yield obtained per saliva specimen) is about 3-fold lower than blood, genomic DNA research requires a very small DNA yield to perform candidate-gene analysis utilizing polymerase chain reaction (PCR) amplification [5]. Saliva samples are a good substitute for blood-derived DNA for individual SNPs with whole saliva DNA purity similar to that of blood extractions [6].
Saliva collection is quick, non-invasive, easy to collect (in-person and by mailed saliva collection kits), and relatively inexpensive method of collecting genetic samples which can be used to simplify the participant requirements for the study [5,7-9]. Study participants will be encouraged to provide a sufficient sample in order to obtain an adequate DNA yield [10].
One problem that has yet to be solved is an economical method of long-term storage of high-quality DNA, which is important for population-based molecular epidemiological studies. In Sigurdson et al., buccal cell DNA was collected from saliva and stored on treated filter cards for seven years at three different temperatures: -20 degrees C, -80 degrees C, and at room temperature. Assessment of the treated cards for buccal cell genome DNA for genotyping after seven years of storage did not consistently provide sufficient quantities of genome DNA [11,12].
While DNA yield is a very important aspect of analyzing saliva collection method, other factors must be taking into consideration. The costs associated with collecting saliva sample include the monetary investment associated with purchasing kits and supplies as well as the resource costs associated with the amount of time required to analyze the genetic material. No matter how great the yield DNA a collection method has, the economically feasibility must be taken into consideration.
Due to the increased acceptance of saliva as a viable alternative to blood samples, the number of commercially available saliva collection kits have appeared on the market. These kits contain stabilization buffers and extraction methods to ensure easy collection methods for research purposes. One popular brand, Oragene, has been studied previously and has been shown to have a DNA yield of approximately 40.4 micrograms per milliliters, with genotyping success in 96% of saliva collections [12]. The purpose of this study is to examine a variety of saliva collection kits and laboratory synthetic/extraction methods and to compare efficiency and yield, without sacrificing quality.
Results and discussion
A DNA yield of above 10 ng/mL was considered high enough for genotyping with PCR extraction. As seen from Figure 2, all but three of the method met this threshold. However, despite no to low DNA detected for Methods 4,7, and 8, all samples were amplified using PCR and were genotyped correctly.
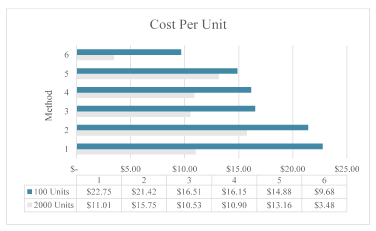
Figure 1. Comparison of cost per unit for methods 1-6 for both 100 units and 2000 units. Methods 7 and 8 were not included in the comparison since they were the same costs as methods 2 and 3
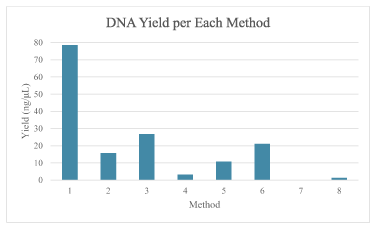
Figure 2. Comparison of average DNA yield for each method. Method 1 had the greatest yield and Method 4 had the least yield
One collection and extraction method examined was outlined in Good, et al. [13] The Goode, et al. paper outlined protocol for creating a buffer, claiming it would yield comparable DNA levels at a fraction of commercially available kits. Goode, et al. was compared to four commercially available saliva collection kits: Oragene Discover, Norgen Biotek, Accegen Aceseq, and Oasis SimplOFy. All kits were sent as sample from the manufacturer. The kits from Oragene and Norgen were used as a whole process; DNA was extracted using the company’s stabilization buffer and extraction method. Accegene and Oasis were examined based on their stabilization buffers.
A second method of DNA extraction was examined with all stabilization buffers. Zymo Quick-DNA Miniprep Plus Kit claimed that it can be utilized to extract DNA from a wide variety of collection methods including different types of samples such as tissue, blood, and saliva utilizing a spin column. The kits are relatively inexpensive, with cost per extraction under six dollars. This method was used to see if the different stabilization buffers would be compatible with extraction methods other than the one developed for the kit.
The least expensive method is Method 6, which paired the stabilization buffer created from the Goode, et al. paper with the Zymo extraction process. At lower purchase order of around 100 extractions, the cost per unit comes out to $9.68, the only method under ten dollars per extraction. At a higher purchase order, economies of scale come into play. With the volume discount offered from Zymo as well as the reduction in costs of purchasing other supplies in bulk (i.e. test tubes and pipets), the cost per unit comes out to $3.48, less than a third of the other methods.
For time comparison, all methods that utilized the Zymo Quick-DNA Midipress extraction methods were completed within half an hour. Norgen Biotek’s process was also around this length. The Oasis and Oragene stabilization buffers required initial incubation prior to extraction which increased time. By far, the longest process was the Goode Method which took almost three hours to complete. This extraction method required the sample be centrifuged multiples and long incubation period. This resulted in a lot of down time waiting for these processes to occur.
All commercially available and lab-synthesized saliva collection kits were capable of yielding genotyping results through PCR amplification. Figure 3 shows the amplification plot of one sample; all samples displayed similar trends. Even samples with DNA yield too low to be measured by the Qubit Fluorometer were able to be amplified and correctly genotyped.
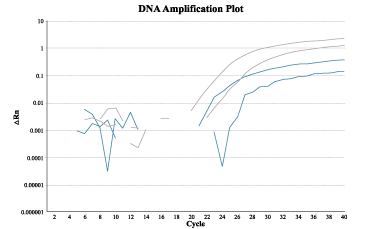
Figure 3. Amplification of DNA after cycles of PCR. All samples displayed similar amplification plots independent of DNA yield quantity
Ease of use comes with the commercial kits as most companies offer extraction processes along with providing kits for collection. For a research project without access to a laboratory, which would be a valid option. However, costs are increased with the purchase of a commercially available kits, with cost per extraction coming to around $10-20 per extraction depending on the bulk amount purchased.
The Goode Extraction Method, while inexpensive compared to its competition, requires a long and arduous extraction process, which might also be a limiting resource. The Zymo Quick-DNA Midipress extraction kit offers an inexpensive solution to the long extraction process of the Goode Method. However, the kit does not come without criticism. Since the kit utilization spin columns, any contaminant in the DNA sample such as undissolved proteins, can clog the filter, reducing DNA yield. When compared to other extraction methods, there is a lower yield in DNA. This method, while less expensive, is more temperamental, and would not be suitable for projects requiring large DNA yields.
There are also criticisms associated with the methods of comparing the saliva collection kits mentioned in this paper. Variation in DNA yield impacted by the amount of DNA in saliva, which can vary by individual. The average amount between two individuals could not be considered as an accurate measurement of the actual DNA yields for each method. It should be noted that even for Method 7, enough DNA was extracted that PCR was able to amplify and genotype correctly.
The buffers required for DNA extraction for the Accengen Aceseq and the Oasis SimplOFy stabilization buffers were not available for this comparison. Therefore, no results were collected regarding these kits for their intended use. The cost of each kit is comparable to their competition in Oragene and Norgen. Therefore, the information for this paper only examined the Accengen and Oasis stabilization buffer in relation to the universal extraction methods of the Zymo Quick-DNA Mediprep Extraction Method.
Finally, the paper did not investigate the long-term effects of DNA extraction and storage between the different methods. All methods claim that the samples could be kept at 4°C storage for up to six months, and up to two years in frozen storage. This was no tested and the long-term stability of the DNA samples could not be inferred.
Table 1. Method, stabilization buffer, and extraction protocols used
Method |
Stabilization Buffer |
Extraction Protocols |
1 |
Goode et al. |
Goode et al. |
2 |
Oragene Discover |
Oragene Discover |
3 |
Norgen Biotek |
Norgen Biotek |
4 |
Acceqen Aceseq |
Zymo Quick-DNA |
5 |
Oasis SimplOFy |
Zymo Quick-DNA |
6 |
Goode et al. |
Zymo Quick-DNA |
7 |
Oragene Discover |
Zymo Quick-DNA |
8 |
Norgen Biotek |
Zymo Quick-DNA |
Table 2. A comparison of Yield, Cost and Time data for the eight Methods
Method |
Stabilization Buffer |
Extraction Method |
Yield (ng/μL) |
Cost: 100 Units |
Cost/Unit |
Cost: 2000 Units |
Cost/Unit |
Time (min) |
1 |
Goode, et al. |
Goode, et al. |
78.45 |
$ 2,274.67 |
$ 22.75 |
$ 22,016.49 |
$ 11.01 |
150 |
2 |
Oragene Discover |
Oragene |
15.65 |
$ 2,141.91 |
$ 21.42 |
$ 31,507.91 |
$ 15.75 |
90 |
3 |
Norgen Biotek |
Norgen |
26.80 |
$ 1,650.56 |
$ 16.51 |
$ 21,065.56 |
$ 10.53 |
20 |
4 |
Accegen Aceseq |
Zymo Quick-DNA |
3.21 |
$ 1,615.00 |
$ 16.15 |
$ 21,800.00 |
$ 10.90 |
20 |
5 |
Oasis SimplOFy |
Zymo Quick-DNA |
10.71 |
$ 1,488.00 |
$ 14.88 |
$ 26,324.00 |
$ 13.16 |
60 |
6 |
Goode, et al. |
Zymo Quick-DNA |
21.15 |
$ 967.61 |
$ 9.68 |
$ 6,966.31 |
$ 3.48 |
30 |
7 |
Oragene Discover |
Zymo Quick-DNA |
- |
$ 2,468.91 |
$ 24.69 |
$ 36,901.91 |
$ 18.45 |
30 |
8 |
Norgen Biotek |
Zymo Quick-DNA |
1.40 |
$ 1,877.56 |
$ 19.78 |
$ 26,459.59 |
$ 13.23 |
30 |
Materials and methods
Five collection methods and two saliva extraction methods were analyzed for a total of eight comparisons, on a variety of metrics to determine efficiency. The eight comparisons were examined in four categories: (1) the DNA yield extracted from each method, (2) the cost per unit for a small purchase order (100 units), (3) the cost per unit for a large purchase order (2,000 units), and (4) total amount of time required to complete the protocol. The eight different methods are a combination of different DNA stabilization buffers, self-made and commercial, and extraction methods.
DNA yield
The comparison of the DNA was done ona small scale. Two volunteers provided saliva samples for each of the collection methods and the DNA yield was averaged between the two. The instructions for each type of collection and extraction method were followed without any deviance.
DNA yields were measured using an Invitrogen Qubit Flurorometer following the company standard assay preparation instructions. The Qubit was re-calibrated with the blank before taking readings of any of the methods. Average DNA yield was recorded as nanograms per microliter. After extracting the DNA, all of the samples were genotyped using PCR method.
Cost
Cost was calculated by examining the materials associated with each extraction method. Quotes were obtained from the respective companies for a small order (100 kits) or a larger order (2,000 kits). Costs associated with additional supplies (i.e. test tubes, tris-EDTA storage buffer) were calculated from commercial scientific research company catalogues. Cost was then divided by the number of extractions to obtain a cost per unit value.
Time
The amount of time was cumulative of the time it took to complete one extraction. This was measured from the start of extraction, including all time required to incubate and centrifuge samples.
Acknowledgment
The authors are thankful to the leadership and staff members of the Center for Pharmacogenomics at The Ohio State University. This work was supported by NIH grant R01 MD.
Author contribution
JS, DW and JPK conceived and designed the experiments. JS performed the experiments and data analysis. JS, DW, SRT and JPK interpreted the analysis results and wrote the article.
References
- Dorak MT (2017) Chapter 8: Candidate Gene Studies and Genome-Wide Association Studies. Genetic Association Studies: Background, Conduct, Analysis, Interpretation. Garland Science 6: 123-162.
- Strachan T, Read Andrew (2011) Chapter 19, Pharmacogenetics, Personalized Medicine and Population Screening. Hum Mol Genet 11: 612.
- Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W (2011) Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J 11: 274-286.
- Snozak CLH and Langman LJ (2012) Chater 1: Pharmacogenomics Principles of Alcohol and DOA. Pharmacogenomics of Alcohol and Drugs of Abuse, Eds Dasgupta, A and Langman, L J, CRC Press ISBN: 9781439856116. 2012: 1-9.
- Gudiseva HV, Hansen M, Gutierrez L, Collins DW, He J, et al. (2016) Saliva DNA quality and genotyping efficiency in a predominantly elderly population. BMC Med Genomics 9: 17. [Crossref]
- Hu Y, Ehli EA, Nelson K, Bohlen K, Lynch C, et al. (2012) Genotyping performance between saliva and blood-derived genomic DNAs on the DMET array: a comparison. PLoS One 7: e33968.
- Loomis SJ, Olson LM, Pasquale LR, Wiggs J, Mirel D, et al. (2010) Feasibility of High-Throughput Genome-Wide Genotyping using DNA from Stored Buccal Cell Samples. Biomark Insights 5: 49-55.
- Nishita DM, Jack LM, McElroy M, McClure JB, Richards J, Swan GE, et al. Clinical trial participant characteristics and saliva and DNA metrics. BMC Med Res Methodol 9:71.
- Etter JF, Neidhart E, Bertrand S, Malafosse A, Bertrand D (2005) Collecting saliva by mail for genetic and cotinine analyses in participants recruited through the Internet. Eur J Epidemiol 20: 833-838.
- Pulford DJ, Mosteller M, Briley JD, Johansson KW, Nelsen AJ, et al. (2013) Saliva sampling in global clinical studies: the impact of low sampling volume on performance of DNA in downstream genotyping experiments. BMC Med Genomics 6: 20.
- Sigurdson AJ, Ha M, Cosentino M, Franklin T, Haque KA, et al. (2006) Long-term storage and recovery of buccal cell DNA from treated cards. Cancer Epidemiol Biomarkers Prev 15: 385-388. [Crossref]
- Rylander-Rudqvist T, Hakansson N, Tybring G, Wolk A (2006) Quality and quantity of saliva DNA obtained from the self-administrated oragene method--a pilot study on the cohort of Swedish men. Cancer Epidemiol Biomarkers Prev 15: 1742-1745.
- Goode MR, Cheong SY, Li N, Ray WC, Bartlett CW (2014) Collection and extraction of saliva DNA for next generation sequencing. J Vis Exp [Crossref]



