Abstract
Introduction: The robustness of the DNA molecule, coupled with the fact that it contains digital information, makes it ideal as a source of identification which resists weather and environmental aggressions imposed on samples found in crime scenes, but these same factors make these samples exhibiting low DNA concentration and with degradation in your chemical structure. The recovery of remaining genetic material of human skeletons is essential part human forensic identification, because the genetic identification by DNA is a method that outperforms all other existing methods.
Objective: This work aims to obtain DNA from bone samples degraded and produce an electropherogram with the minimum number of markers recommended by the scientific community forensic, using a simple and effective protocol.
Materials and methods: In order to analyse the DNA, samples of the bones and the teeth was demineralized in an EDTA solution and extracted using the Microcon® tube protocol. PCR analysis system approached by STRs PowerPlex Fusion 6C® system, PowerPlex Fusion® system and Globalfiler® System and Capillary electrophoresis procedures were performed on the ABI 3500 Genetic Analyzer.
Results: The DNA extracted from the teeth sample and from the bone sample with demineralization without pulverization presented a male genetic profile complete with all amplification system markers.
Conclusions: Our alternative extraction protocol proved to be efficient because it allowed the isolation of DNA from high quality of a 17-year bone buried, in order to present a complete genetic profile of all system markers.
Key words
forensic DNA, bones DNA, bones decalcification
Introduction
One of the biggest challenges to public security and judicial area has been the identification of corpses in advance state of decomposition or skeletonization and it has generated a lot of anguish for the professionals and, above all, to the relatives of these victims, who lost their relatives and are entitled to answers of the governmental entity.
In corpses, whose time of death is more than two years, the biological material of choice are the bones, soft tissues no longer exist, the teeth get lost easily and the hairs, if any, still have meagre quantities and degraded DNA. Already the extraction of hard tissues such as bones, provide results that are more satisfactory, because they become a natural deposit of nucleic acids protected the environment. However, after the death, is the balancing of the repair process, which leads to progressive and cumulative damage, which is why nucleic acids will gradually be being degraded through hydrolysis and oxidation processes [1].
The steps of extraction and purification of DNA in bones have been the most difficult and complicated for human identification through this fabric in forensic laboratories. The lack and degradation of the DNA of bones buried or abandoned in various terrains get along by contact with fungi, bacteria, humic acids and fulvic acids, which are abundant in soils and can be coextracted along with human DNA, causing the inhibition of the PCR reaction.
Due to that fact, are spent days or sometimes weeks in extraction process of DNA, unlike analyses carried out in laboratories that only work with paternity investigation in alive, where the material used is blood or saliva, that presents itself in large quantity and in great condition [2].
In order to succeed in obtaining DNA from bone samples degraded and produce an electropherogram with the minimum amount of recommended by the scientific community forensic markers, it is imperative that the most modern techniques of extraction and DNA purification.
One of the methods commonly used by forensic laboratories for sample preparation, are the methods of cutting and sanding of these samples, or the use of liquid nitrogen and cryogenic mills for pulverization of the samples, in order to turn the powdered bone. Some laboratories also use automated ways for extracting DNA from bones, but these are rather expensive, considering that require the purchase of the equipment supplied by specialized company [3].
Descaling is based on inorganic phase destruction of bone tissue, to calcium removal, that will get to the organic portion of the bone. To this end, the techniques more usually employed are those, which use strong acids, which form soluble calcium salts and chelating agents, who kidnap the calcium ions leaving the organic portion more pliable and easy removal [4].
One of the acids with decalcifying properties, the most used in these protocols are nitric acid, hydrochloric acid, formic acid and some other hydrochlorides acids. Although these acids decalcify the bone quickly, cannot be used for extended period, because they cause the acid hydrolysis of DNA, compromising your stability [5]. In experimental studies, Sarsfield et al. [6] found that the formic acid degrades the DNA, since no DNA was previewed on electrophoresis, after using the acid in the samples referred to.
Currently, most of the decalcification protocols in the area of Genetics and Histology is based on the use of EDTA (Tetra-acetic ethylenediamine acid), which is a low molecular weight organic acid, which demineralize bone and inactivates DNAses enzymes, by chelating bivalent cations like the MG ++ and Ca ++. The decalcification by EDTA has been shown to be the best technique of recovering DNA from bone tissue, when compared to other techniques that use other acids, because EDTA has high ability to remove calcium ions from bone matrix by abstracting the crystals formed [7].
EDTA acts only in the mineralized bone matrix, not committing any other fabric, especially the genetic material contained in the nucleus of cells. Due to that fact, are preferable to use techniques that descaling agent to those that use other descaling agents that modify or degrade the DNA, and therefore unsuitable for analyses that are intended to obtain DNA [5].
After this pre-treatment, organic extraction is performed, in which are used organic solvents, like phenol, associated with chloroform. This method removes the proteins attached to DNA, causing this has better quality and purity [8].
After this pre-treatment of demineralization, organic extraction is performed, in which uses organic solvents, like phenol, associated to chloroform, with subsequent centrifugation and filtration in silica columns. This method removes the proteins attached to DNA, causing this has better quality and purity [8].
According to Budowle et al. [9], the ideal extraction protocol is that for the vast majority of the samples is of simple implementation and has low cost for the lab.
Whereas in forensics, is the imminent need for obtaining DNA from bones and teeth by using simple and effective protocols, the present work aims at the development of alternative protocol for demineralization without pulverization at extraction and purification of DNA from ancient human forensic samples and degraded for forensic identification [10].
Materials and methods
The analysis of biological samples in the context of Forensic Genetics, both in forensic biological affiliation and identification of bodies and human remains is developed in two phases: laboratory testing and biostatistics analysis of results, having been analyzed in this study, the following samples:
For development of alternative protocol of demineralization and DNA extraction of ancient bones degraded, 08 were analyzed samples of 01 fragment of a femur of corpse (named MTS), that was buried for 17 years. As reference sample, we used 02 samples of 04 teeth of corpse, which was buried for 3 years (named MS), being this, supposed biological son of corpse buried for 17 years.
A fragment of the femur, 2 cm by 3.7 cm in diameter, was cleaned with ultra-pure water and gauze and then incubated in 20 ml of ethylenediamine acid tetra-acetic- solution – EDTA 0.5M, pH 8, by 30 days at room temperature – between 25 and 30ºC. The four teeth were also incubated in the same solution, but for only 15 days, also at room temperature.
After demineralization, the fragment of the femur was washed with ultra-pure water and clean with gauze, after that, were remove, with the aid of a scalpel, splinters of demineralized femur fragment, having been placed about of 0.3 g of these splinters in eight micro tubes capacity 1.5 mL Eppendorf type. Similarly, 0.3 g of the splinters of the demineralized teeth were placed in two other micro tubes of same capacity. Each of the sample, were added 600 µL of extraction buffer (10 mM Tris-HCl, 100 mM NaCl; 10 mM EDTA; 2% SDS, pH 8.0), 60 µL of Proteinase K and 40 µL of DTT, having been incubated overnight these to 56°C.
After the incubation period, 600 µL were added phenol/chloroform/isoamylic alcohol (25/24/1, v/v/v) in each of the samples, which were subject to unrest in vortex, until the formation of a milky solution. Then, the samples were subjected to centrifugation for 7 minutes at 12,000 rpm, for the formation of the supernatant (aqueous phase) separating the DNA of proteins. About 500 µL of the supernatant of each sample was transferred to a membrane filter tube with silica Microcon® brand.
All 10 samples were centrifuged for 9 minutes at 12,000 rpm and the filtered liquid was disposed one by one, being the filter with the DNA, reattached the tube. After that were added in the filter, 500 µL of ultra-pure water for washing of the DNA, and then centrifuged for 9 minutes at 12,000 rpm. The filtered liquid was dropped and added again, two more times, 500 µL of ultra-pure water for washing. Recovery of DNA joined the silica membrane was made with the addition of 50 µL ultra-pure water, with the reversal of the filter on a new Microcon® tube.
The polymerase chain reaction (PCR) of all samples were conducted in thermal cycler Proflex brand, the company Life Technologies for amplification cycles 29, with two phases, the first being with two stages and the second, with only one stage.
The samples analyzed i2021 Copyright OAT. All rights reserv genotyped for STRs markers, which are the most commonly used by the forensic community.
The DNA extracted from teeth samples was submitted to amplification, using the commercial multiplex kit PowerPlex Fusion 6C of the company Promega Corporation, which owns 27 markers. The amplification product of this sample was genotyped to the loci D1S1656, D3S1358, D2S441, D10S1248, D13S317, D16S539, D18S51, Penta E, D2S1338, CSF1PO, Penta D, TH01, D21S11, D7S820, vWA, TPOX, D5S818, D8S1179, D12S391, D19S433, D22S1045, SE33 DYS391, FGA, DYS576 and DYS570, besides the amelogenin, to identify the gender.
The DNA extracted from the bone samples were submitted to amplification, using the commercial multiplex PowerPlex Fusion kit the company Promega Corporation, which has 25 markers, not possessing just two markers present in the commercial kit PowerPlex Fusion 6C (DYS576, DYS570). The amplification products of these samples were genotyped to the loci D1S1656, D3S1358, D2S441, D10S1248, D13S317, D16S539, D18S51, Penta E, D2S1338, CSF1PO, Penta D, TH01, D21S11, D7S820, vWA, TPOX, D5S818, DYS391, D8S1179, D12S391, D19S433, SE33, D22S1045, FGA, in addition amelogenin, to gender identification.
The DNA extracted from the bone samples were also submitted to amplification, using the commercial multiplex kit GlobalFiler® of company Life Technologies, which has 24 markers. The amplification product of this sample was genotyped to the loci D3S1358, vWA, TPOX, D16S539, CSF1PO, D8S1179, D21S11, D18S51, DYS391, D2S441, D19S433, FGA, TH01, D22S1045, D5S818, D13S317, D7S820, D10S1248, D1S1656, SE33, D2S1338, D12S391 and Y indel, beyond amelogenin, for gender identification.
Capillary electrophoresis procedures were performed on sequencing platform ABI 3500 Genetic Analyzer, with POP-4 polymer and 36 cm capillary (Applied Biosystems).
According to Arenas et al. [10] the identification of individuals can be obtained through a number of sufficient markers to enable a high power of discrimination, and it is important to check the quality of the profiles before analysing the results. After that, the genetic information obtained are compared with other database information or reference samples, performing the statistical analysis.
To establish the link between the samples is calculated the likelihood ratio - LR, which is the ratio of the probability of obtaining an identity of genetic profiles if the DNA in the sample questioned and suspect come from the same person, and the probability of a DNA come from different people. The LR is the reciprocal of the probability of random combination and a value of 1000 means that is 1000 times more likely to get an identity, if DNA samples are from the same person, than if they came from two individuals any chosen randomly in the population [11].
According to Arenas et al. [10] for identification of human beings, are used preferentially the autosomal STRs, which are analyzed with a Bayesian approach (prior probabilities are combined with probabilities of genotype observations, assuming alternative hypotheses).
In this work, for analysis of the results of the samples of bones and teeth, was used the statistical program FAMILIAS, version 3.2.1, which analyses the index of parental relations according to allele frequencies reported of shared alleles.
Statistical calculations were tested two probabilities: 1) the body of MS is a biological child of the corpse of MTS; 2) the body of MS is a biological child of another person chosen at random in the Brazilian population.
Results
The DNA extracted from the teeth sample with demineralization without pulverization presented a male genetic profile complete with all 27 amplification system markers PowerPlex Fusion 6C of the company Promega Corporation, with 23 markers autosomal, 03 male line markers and marker for the identification of gender – amelogenin (Figure 1).
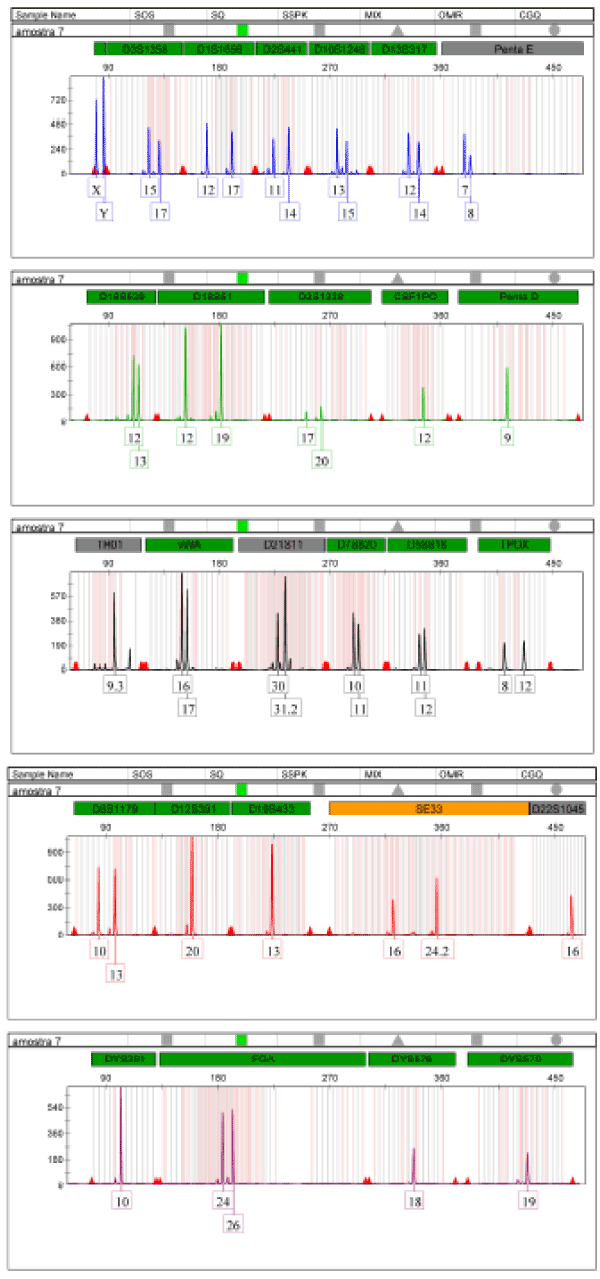
Figure 1. Electrophoresis of DNA amplification extracted from the teeth sample with the PowerPlex Fusion 6C system from Promega Corporation
The DNA extracted from the sample of bone with demineralization without pulverization presented a male genetic profile complete with amplification of all 25 markers PowerPlex Fusion system of company Promega Corporation, and autosomal markers 23, 01 male line marker and the marker for the identification of gender – amelogenin amelogenin, having been found a triallelic pattern on TH01 (Figure 2).
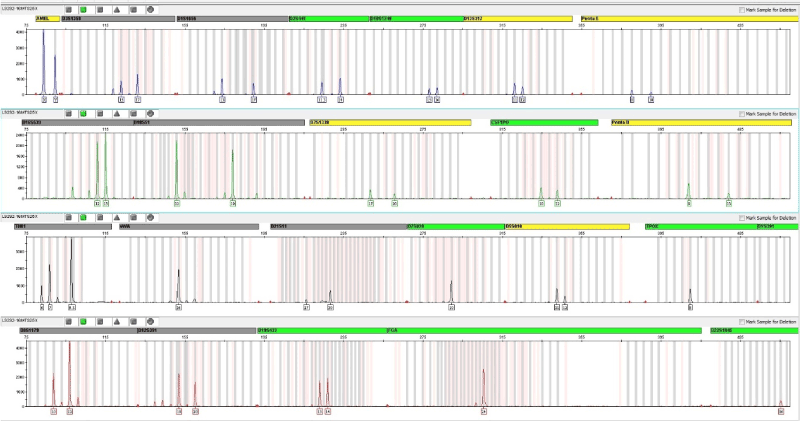
Figure 2. Electrophoresis of amplification of DNA extracted from the bone sample with the PowerPlex Fusion system from Promega Corporation
During the analysis this case, it was observed a tri- allelic pattern type 1 in the locus TH01, with 6, 7 and 9.3 alleles. The TH01 marker is part of the 20 loci STR panel of the Combined DNA Index System (CODIS). There are no articles reporting the occurrence of trialelia in locus TH01. The NIST STRBase database contains described 7 cases of tri-allelic patterns for TH01 locus. The tri-allelic pattern most frequent to the locus TH01 is (7, 8 and 9), having been found in a total of 3 cases, in the absence of any description with the pattern (6, 7 and 9.3). This data is available in the database of STRs: <https: strbase.nist.gov/tri_tab.htm> and are updated often by the National Institute of Justice of the United States of America.
The DNA extracted from the sample of bone with demineralization without pulverization presented a male genetic profile complete with all 24-amplification system GlobalFiler® markers of company Life Technologies, being autosomal 21, 01 to lineage male, male sexual confirmation 01 (Y indel), besides the amelogenin, for identifying the gender (Figure 3).
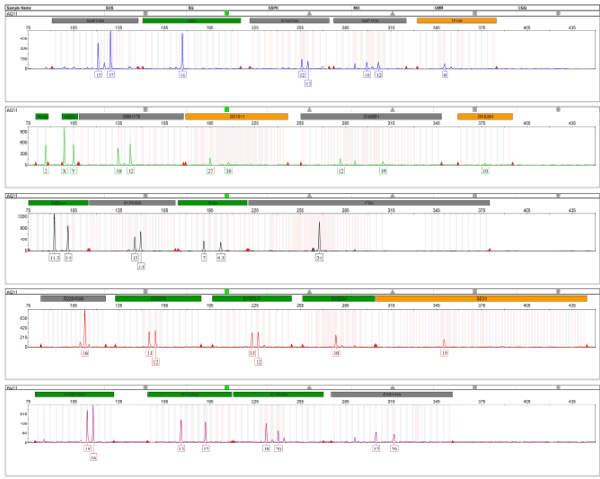
Figure 3. Electrophoresis of amplification of DNA extracted from the bone sample, with GlobalFiler® system from Life Technologies.
In the evaluation of the results of Forensic Genetics, statistical analysis shows a predominant role, because it is necessary to evaluate how particular individual may or may not be related to certain genetic evidence.
In this sense, the estimative of statistical parameters forensics aims to ascertain whether the genetic markers used in forensic genetics are discriminatory and ultimately to calculate with great credibility which the probability of an individual being related to given process (criminal or civil) compared to another individual in the population.
It is important to stress that these are comparative analysis, by means of which will be inferred conclusions based on joint analysis of reference samples and questioned samples.
The study of nuclear DNA polymorphisms of the autosomal chromosomes, in this study allowed determining genetic profiles of two males; where there is sharing of alleles at all loci analyzed bone samples of MTS and of the teeth sample of MS, establishing paternity inclusion of that condition in relation to this.
Once the genetic link compatibility and based on the values of allele frequencies published for the Brazilian population has made the statistical calculations using the statistical program 3.2.1 version FAMILIES, aiming to estimate the weight of the evidence.
Statistical analysis of genetic profiles in question allowed estimate is 23,061,412 times more likely that the body of MS is a biological child of the corpse of MTS, than another person chosen randomly in the Brazilian population, with probability of 99.99999%.
The traditional protocols of extracting DNA from bones and teeth through the traditional organic method, establish the demineralization of these materials prior to incubation in a solution of 0,5 m EDTA, pH 8.0, for a period of five days for up to 2 years of bone death. For these materials with smaller postmortem interval, uses the traditional organic extraction and subsequent amplification and genotyping of type STR markers, which allow to discriminate against an individual in genetically a gene pool of a trillion people.
For bones and teeth with more than 2 years of death, is also recommended prior demineralization, but most studies show that satisfactory results of genetic identification only been obtained through DNA sequencing mitochondrial, which presents itself less degraded in ancient bones and teeth.
Moreti and Tiago [1], assessed nine extraction methods of DNA from bones in 19 bones of 07 individuals, of whom, 02 were originate from exhumation; 03 came from ground deposition of earth; and 02 came from aquatic environment. Of the nine methods evaluated, eight were processed with the spraying of the fragments of the samples and only one, rendered with the fragments of the intact samples (not pulverized), having obtained DNA from almost all the samples, however, most of the DNA detected not was intact. Success was obtained in extraction of DNA from only a sample not pulverized (body deposed in soil of earth), for which there was amplification and genotyping of 9 loci autosomal STRs more sexual locus in the electropherogram.
Carvalho [12] evaluated two methods of extracting DNA from bones in six samples of femur of two human bones exposed for a period of three months to various environmental conditions, such as heat, rain and humidity. The samples processed by the organic method, with prior descaling without spraying resulted in amplification and genotyping of 15 loci autosomal STRs more sexual locus in the electropherogram. The samples were submitted for DNA extraction at traditional organic method (with pulverization) does not result in obtaining DNA that allow amplification of any loci in the electropherogram, not generating a genetic profile.
Hasan et al. [3] analysed 52 samples of remaining bones of bodies of victims of a building collapse in Dhaka, collected in the period from 7 to 15 days after the disaster. These samples were incubated without spraying in EDTA solution (0, 5 m, pH 7.5), for 15 days, being the solution changed every 24 hours. After PCR and genotyping of extracted DNA, most of the samples presented on 15 STR loci full profile, plus amelogenin.
Loreille et al. [13] analysed the efficiency of extraction of DNA from bones sprayed with total demineralization and obtained good results with just the sequencing of mtDNA and with Low Copy Number analysis, also suitable for samples containing little DNA. Another study of Adler et al., also made use of mtDNA sequencing to identify human bones, with powdering, obtaining satisfactory results.
Other studies that used STRs markers for old bones and teeth, showed amplification and genotyping of alleles in up to 15 bullets, as verified [3,4].
Conclusions
In some cases, modified versions of these protocols, or additional steps are required to obtain a satisfactory result, for this reason, our alternative extraction protocol proved to be efficient because it allowed the isolation of DNA from high quality of a 17-year bone buried, in order to present a complete genetic profile of 27 markers, including the sexual marker (amelogenin), with the PowerPlex system Fusion 6C of the company Promega Corporation and a complete genetic profile of 24 markers, including sexual (amelogenin) marker, and the Y Indel marker, with the GlobalFiler® system of company Life Technologies.
Acknowledgements
To Laboratório Forense of the Perícia Oficial de Alagoas for the collaboration and the support given to this study. All funding was provided by Government of the State of Alagoas, Brazil.
References
- Tiago M (2009) Uma proposta metodológica para obtenção de DNA de ossos e implementação de banco de dados de frequências alélicas de STRs autossômicos na população de Santa Catarina. Dissertação de Mestrado em Biotecnologia, Universidade Federal de Santa Catarina. Florianópolis.
- Molly Fitzgerald-Hayes, Reichsman Frieda (2014) DNA and Biotechnology. 3rd edn. San Diego: Elsevier.
- Hasan M, Hossain T, Majumder AK, Momtaz P, Sharmin T, et al. (2014) An efficient DNA extraction method from bone and tooth samples by complete demineralization followed by the use of silica‐based columns. Dhaka Univ J Biol Sci 23: 101‐107.
- Balayan A, Kapoor A, Chaudhary G, Raina A (2015) Evaluation of techniques for human bone decalcification and amplification using sixteen STR markers. Egyptian Journal of Forensic Sciences 5: 30-35.
- Singh VM, Salunga RC, Huang VJ, Tran Y, Erlander M, et al. (2013) Analysis of the effects of various decalcification agents on the quantity and quality of nucleic acid (DNA and RNA) recovered from bone biopsies. Ann Diagn Pathol 17: 322-326 [Crossref]
- Wickham CL, Sarsfield P, Joyner MV, Jones DB, Ellard S, et al. (2000) Formic acid decalcification of bones marrow degrades DNA: alternative use of EDTA allows the amplification and sequencing of relatively long PCR products. Mol Pathol 53: 336-337. [Crossref]
- Balayan A, Kapoor A, Chaudhary G, Raina A (2015) Evaluation of techniques for human bone decalcification and amplification using sixteen STR markers. Egyptian Journal of Forensic Sciences 5: 30-35.
- Butler JM (2005) Forensic DNA Typing. 2ndEdn. London: Elsevier Academic Press.
- Budowle B, Brown BL (2001) The use of DNA typing for forensic identification. Forénsica 1: 23-37. 2001.
- Arenas M, Pereira F, Oliveira M, Pinto N, Lopes AM, et al. (2017) Forensic genetics and genomics: Much more than just a human affair. PLoS Genet 13: e1006960. [Crossref]
- Ana Cláudia P (2010) Emprego de mini-STRs “non-CODIS” em amostras biológicas de DNA forense. Dissertação de Mestrado em Biotecnologia, Universidade Federal de São Paulo. São Paulo.
- Hérika GAC (2009) Extração de DNA de ossos humano, sem pulverização, para uso em identificação forense. Dissertação de Mestrado em Ciências Biológicas, Universidade Federal de Pernambuco. Recife.
- Loreille OM, Diegoli TM, Irwin JA, Coble MD, Parsons TJ (2007) High efficiency DNA extraction from bone by total demineralization. Forensic Sci Int Genet 1: 191-195. [Crossref]



