Abstract
In the reductive pyrimidine catalytic pathway uracil and thymine are converted to β- alanine and β-aminoisobutyrate. The amidohydrolases of this pathway are responsible for both the ring opening of dihydrouracil and dihydrothymine and the hydrolysis of N-carbamoyl-β-alanine and N-carbamoyl-β-aminoisobutyrate. The review summarize the properties, kinetic parameters, 3D-structures and reaction mechanisms of these proteins. The two amidohydrolases have unrelated folds, the DHPases belong to the amidohydrolase superfamily, while the βAS from higher eukaryotes belongs to the nitrilase superfamily. βAS from S. kluyveri is an exception to the rule and belongs to the Acyl/M20 family.
Key words
dihydropyrimidine amidohydrolase, β-alanine synthase, hydantoinase, amidohydrolase superfamily, nitrilase superfamily, 3d-structures, amino acid triad, β-alanine aminotransferase
Abbreviations
βAS: β-alanine synthase; HβAS: human β-alanine synthase; NCβA: N-carbamoyl-β-alanine; DmβAS: β-alanine synthase from D. melanogaster; βAIBA: β-aminoisobutyrate; 5FU: 5-fluorouracil; DHPase: dihydropyrimidinase
In the catalytic pathway of pyrimidines, uracil and thymine are reduced to 5,6-dihydro-Uracil and 5,6-dihydrothymine, respectively, by dimeric dihydropyrimidine dehydrogenase (DPD) (EC 1.3.1.2.), a flavoenzyme containing 4 iron sulphur clusters [1]. The second reaction catalysed by 5,6-dihydropyrimidinase (DHPase) (EC 3.5.2.2.) is the reversible ring opening between N3 and C4 of the pyrimidine ring, resulting in N-carbamoyl–β-alanine and N-carbamoyl-β-aminoisobutyrate, respectively. In the third step,the N -carbamoyl group by β-alanine synthase) βAS) (EC 3.5.1.6) to form β-alanine and β-amino-isobutyrate, respectively.The carbamoyl group decomposes to carbon dioxide and ammonia [2]. In some eukaryotes, the transamination of β-alanine in the presence of α-ketoglutarate to malonate semialdehyde and glutamate by β-alanine aminotransferase (EC 2.6.1.19) is taking place [3] (Scheme 1).
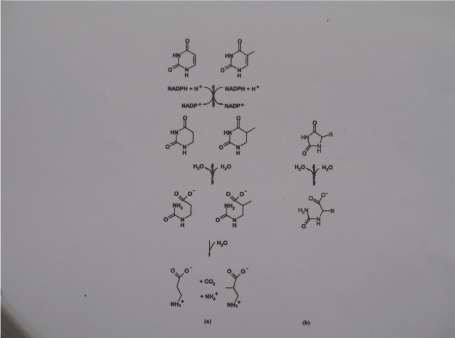
Scheme 1. a) the reductive degradation pathway for uracil and thymine. b) hydrolysis of 5-)monosubstituted hydantoin as catalysed by hydantoinases
In mammals, β-alanine can only be produced by reductive degradation of uracil [4]. β-Alanine is widely distributed in the central nervous system in form of β-alanine containing dipeptides such as anserine, carnosine and (β-alanyl(ornithine) [5]. In addition it is a structural analogy of gamma-aminobutyrate )GABA) and glycine to major inhibitory neurotransmitters [6]. The reductive pyrimidine catabolic pathway is also the main clearance route for cytotoxic pyrimidine analogues such as 5-fluorouracil (5FU) [7] and has therefore important medical implications. 5FU is still the most widely used anti-cancer therapeutic [8]. The pyrimidine catabolic pathway, however, degrades most of the administered 5FU in a short period [8], i.e., only a few 5FU molecules reach the ultimate target, thymidylate synthase. Large amounts of 5FU have to be administered with the consequence of increase toxicity of the 5FU treatment. This review summarizes the biochemical, structural and mechanistic data available for amidohydrolases.
Amidohydolases of the pyrimidine degradative pathway
Purification, properties, substrate specificities, and sequence homologies of dihydro-pyrimidinases
Dihydropyrimidinases (DHPases) from bovine, calf and rat liver [9-15] as well as plants and fungi [16,17] have been purified from natural source or as recombinant proteins.
All characterized DHPases are homotetrameric zinc-metaloenzymes,the molecular masses of DHPase subunits range from 56 kDa for slime mould species to 65 kDa for the fruit fly enzyme [18]. Mammalien DHPases use not only DHU and DHT as substrates but also catalyse the hydrolysis of various 5-monosubstituted hydantoins and succinimides [19]. But not all hydantoinases, the bacterial counterparts of DHPases hydrolyse 5,6-dihydro-pyrimidines [20,21].
DHPases exhibit 24-43% amino acid sequence identity to hydantoinases, whereas pairwise sequence identities between DHPases range from27% for the evolutionary distant species (human/rat) to 94% for close DHPase relatives (rat/mouse).
Enzymatic activities in the forward direction can readily be observed at pH 8.0 by following the disappearance of the characteristic absorbance of DHU and DHT at 225 nm [22]. The pH-independent V/KDHU for the pig liver enzyme is 1.29x10-2 M-1 s-1. The reverse reaction is optimal at pH 5.5. Inhibitors of DHPases are products, N-carbamoyl-β-alanine (Ki = 0,68 mM), glutarate monoamide (Ki = 0.21 mM), And 4-ureidobutyrate (Ki = 0.99 mM)
Dihydropyrimidinase deficiency
Dihydropyrimidinase deficiency is a rare event that causes dihydropyrimidinuria. Patients may be largely asymptomatic or show a variable clinical phenotype comprising seizures, mental retardation, growth retardation, and dysmorphic features [23]. Furthermore, dihydropyrimidinase deficiency in humans has been associated with
increased risk of 5FU toxicity [23,24].
Purification, properties, substrate specificities and sequence homologies of β-alanine synthases
β-Alanine synthase: βAS from calf, human, rat liver [22] from Pseudomonas putida [23] as well as from plants and fungi [16,17]. All characterized βAS are either homodimers (human, S. kluyveri), homohexameric (calf, rat liver) or homododecameric (Arabidopsis thaliana,Zea mays) enzymes. The molecular masses of βAS range from 40 kDa for the calfliver enzyme to 50 kDa for the SKβAS species [18].
Enzymatic activities were either determined by derivatizing the formed β-alanine with phenyl isocyanate followed by HPLC chromatography of the PTC-β-alanine [22], or using radioactive 2-C14-labeled N-carbamoyl-β-alanine and determination of radioactive carbon dioxide [25].
Inhibitors of the calf liver βAS are 4-ureidobutyrate, propionate and glutarate monoamide with Ki values of 1.6, 240, 630 µM, respectively [22]. Furthermore, kinetic studies have shown that rat βAS is an allosteric protein with positive co-operativity towards N-carbamoyl-β-alanine that triggers a change in the oligomeric state from homohexamer to homododecamer, whereas the presence of β-alanine induces the dissociation into inactive trimers [26]. This behaviour was not observed in the calf liver enzyme. A reason for the difference in behaviour could be the fact that in the purification of calf liver βAS a heat step under the protection of propionate was used which may be responsible for the desensitisation of the calf liver enzyme [22].
β-Alanine synthase deficiencies
The frequency of β-alanine synthase deficiency appears to be rather low. The first case of such enzyme deficiency was reported in 1999 [27]. An 11 months old girl with severe developmental delay and dystonic movement disorder, showed abnormally high amounts of β-ureidopropionate and β-ureidoisibutyrate in the urine. Other patients with a defect β-alanine synthase suffering of neurological disorders have been reported [28].
Three-dimensional Structures
Dihydropyrimidine amidohydrolases
Amidohydrolases are a large group of proteins which catalyse the hydrolysis of a wide range of substrates bearing amide and ester functional groups at carbon or phosphorus centres. Concerning their 3D-structures, these enzymes can be separated into different fold families. The two amidohydrolases of the pyrimidine catalytic pathway have different folds, with DHPases belonging to the amidohydrolase superfamily [29] while the β-alanine synthase either belongs to the nitrilase [30] or the Acyl/M20 family of the metapeptidases [31] depending on the source of the enzyme. Raushel and coworkers have suggested that most outstanding structural landmark for the amidohydrolase superfamily is a mononuclear or binuclear metal centre embedded with the a (a/β)8 structural fold [32].
The fold of the homodimeric βAS from S. kluyveri is a member of the Acyl/M20 family of metallopeptidases [31, 33], which consist of one or two domains with a/β-fold. In contrast, βASs from higher eukaryotes are expected to show structural similarity to N-carbamoyl-D-amino acid amidohydrolases and other members of the nitirlase
superfamily (Figure 1).
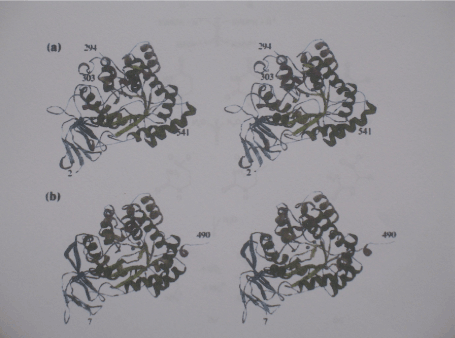
Figure 1. The subunit structure of DHPase
- a) Stereoview of the subunit of DHPase from S. kluyveri. The following colour Coding is used for the secondary structure elements: green and dark green, barrel Strands and helices , respectively; blue and light blue, strands of the larger and smaller Sheet of the ß-sandwich domain, respectively; yellow, additional ß-strands; orange and Dark orange, additional a- and 310 helices, respectively. Spheres in magenta represent the zinc ions of the metal centre. b) Stereo view of the subunit of DHPase from D. discoideum. The same colour coding as used in a) centre
In recent years, structural information has been reported for DHPase-homologous amidohydrolases, e.g. urease [34], dihydroorotase [32] and four hydantoinases with D- and one with L-stereoselectivity [35-37].
Subunit structure of dihydopyrimidine amidohydrolase: For most members of the amidohydrolase superfamily [38] the subunit of DHPases can be divided into two domains, a core catalytic domain and a smaller β-sandwich domain.
Di-metal centre of dihydropyrimidine amidohydrolases: The DHPase active centre is confined by the C-terminal ends of the barrel strands and harbors a dinuclear zinc centre (Figure 2). According to the classification of structurally characterized metal centres within the amidohydrolase super family, it belongs to subtype 1. The geometry of the di-zinc centre and the type and position of amino acid side chains involved in its ligation are strictly conserved between DHPase from S: kluyveri and D. discoideum [39]. The close proximity of the positively charged metal ions makes it likely to be deprotonated to a hydroxyl ion. A second bridge is provided by the carbamoyl group of the carboxylated lysine residue (Lys167 for the S. kluyveri DHPase and Lys158 for D. melanogaster enzyme) The other ligands of Zn-α are an aspartate (Asp358 and Asp325 for the S. kluyveri and D. discodeum species, respectively and two histidines (His62 (His66)) and His64 (His68), respectively. Two additional histidins (His199 or His191 and His225 and His247, respectively) coordinate Zn-β. The pent a coordination of Zn-alpha has distorted trigonal bipyramidal geometry with Lys and Asp as apical ligands, whereas the geometry of the Zn-β coordination is tetrahedral distorted (Figure 3).
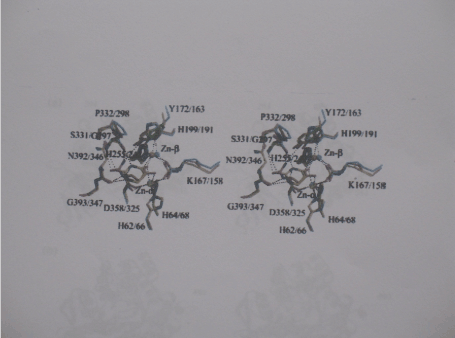
Figure 2. Stereo view of the di-zinc centers and ligand binding sites of DHPase
Superposition of the ligand-binding sites of DHPase from S. kluyveri incomplex with the Substrate DHU (yellow carbon atoms), DHPase from S, kluyveri with the product NCßA (orange carbon atom), and ligand-free DHPase from D. discoideum (cyan carbon atoms). All residues are labelled with the first number corresponding the sequence of DHPase From S. kluyveri. Zinc ions are represented as spheres, the coordination, the zinc ions and hydrogen bonding interactions to ligands are indicated as dashed lines
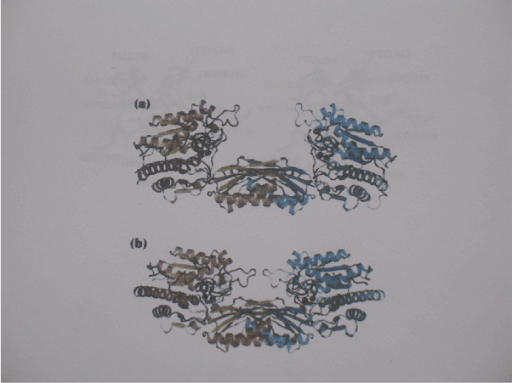
Figure 3. Open and closed conformations of ßAS from S. kluyveri
- a) Schematic view of the extended open enzyme conformation as observed for ligand-Free ßAS from kluyveri. The two subunits are coloured differently. The zinc ions of the di-metal centers are shown in black spheres. b) Schematic view of the homodimer of ßAS-E159A from S. kluyveri observed in the closed conformation. Substrate molecules bound in the respective active site are shown as ball-in-stick models with carbon atoms in white, oxygens in red and nitrogen atoms in blue
Substrate binding of dihydropyrimidine amidohydrolases: Soaking of DHPase from S. kluyveri crystals prior to data collection in a mother liquor-like solution of altered pH, containing both substrate DHU and the product NCβA led to appearance of additional electron density in the active sites of all four protomers. The electron density can clearly attributed to a bound DHU molecule. Only two amino acid residues are directly involved in substrate binding. The backbone carbonyl of Asn392 forms a hydrogen bond with the N1 of the dihydropyrimidine ring. Ser331 forms two hydrogen bonds with the substrate; its amide nitrogen interacts with O-2, and backbone oxygen of the substrate interacts with N-3 of DHU. The remaining ring oxygen of the substrate points towards the binuclear metal centre and is about 2 Angstrom from Zn-β and 2.4 Angstrom from Zn-a [39] (Figure 4).
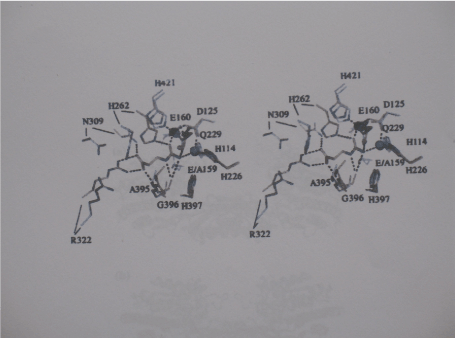
Figure 4. Substrate binding to ßAS from S. kluyveri Stereoview of the superimposed active sites of the complex of ßAS-E159A with the Substrate NCßA (closed state, carbon atoms (orange) and of the ligand-free, wild-type Enzyme (open sate, carbon atoms in white). Zinc ions in blue belong to the latter Structure, zinc ions in black of the ßAS
Active Site of DHPase from D. discoideum: Comparison of the active site of DHPase from D. discoideum with that of the yeast enzyme reveals that the vast majority of the amino acid side chains with a radius of 8 Angstrom from the substrate molecule are identical. The substitution of Ser331 by a glycine in DHPase from D.discoideum should not affect substrate binding, because only main chain atoms of the residue interact with the ligand.
Reaction mechanism of dihydropyrimidine amidohydrolases: Based on the high level of active site conservation to dihydrootase (DHOase) and the pH-dependence of kinetic parameters and solvent deuterium isotope effects there is a general agreement that enzymatic catalysis in hydantoinases and DHPases follows a mechanism very similar to to that for DHOase [32,40]. According to this mechanism, a water molecule in the coordination sphere of the more solvent exposed Zn-β is replaced by the oxo group of the incoming substrate. The interaction polarizes the carbonyl group and prepares the carbon of the scissible bond for hydrolysis (Figure 5).
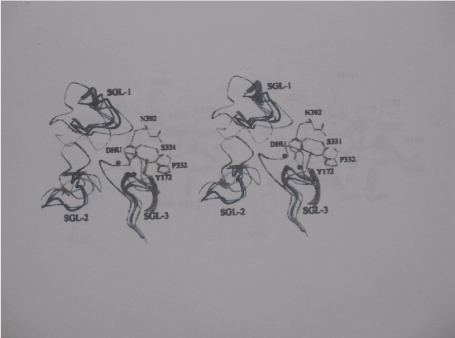
Figure 5. Stereochemistry gate loops in DHPases and hydantoinases. The superimposed backbones of the SGLs as founding the DHPase-DHU complex from S. kluyveri (yellow), in DHPase from D. discoideum (orange), DHPase from B9 (blue), DHPase from Bs (dark Blue), DHPase from Bp (cyan), L-hydantoinase (green) are Shown. The location of the active site is indicated by ball-in-stick representation of The two zinc ions (black spheres), DHU, substrate-binding residues, and Tyr172 (all with carbon atoms in yellow) for the DHPase-DHU complex from S. kluyveri
Comparison to other DHPases and Hydantoinases,
Overall Structure
A sequence alignment of DHPases and structurally characterized hydantoinases reveal that DHPase from S. kluyveri contains several insertions that are not found in the other sequences. A biological function of inserted stretches is not clearly evident, as all located on the periphery far from the active site [39]. Compared with hydantoinases of known structure, the C-termini of all known DHPase sequences are extended. The extension is 18, 38, 110 amino acids long for DHPases from S. kluyveri, D. discoideum and D. melanogaster, respectively [39].
β-alanine synthases
β-Alanine synthase is also called ureidopropionase and N-carbamoyl-β-alanine amidohydrolase (EC 3.6.1.6), It catalyses the irreversible last step in the reductive degradation of the pyrimidine nucleotide bases uracil and thymine, the hydrolysis of N-carbamoyl-β-alanine and N-carbamoyl-β aminoisobutyrate to β-alanine and β-amino-isobutyrate respectively, under the release of carbon dioxide and ammonia [41].
Three-dimensional structure of β-alanine synthases: The structure of DHPase from S, kluyveri has been determined to 2.7 Angstrom resolution by the multiwavelength anomalous dispersion method in two different crystal forms, both belonging to the monoclinic space group P21 but exhibiting distinct unit cell dimensions [31]. Two Zn2+ ions are bound in the active site of each subunit. Human ureidopropionase crystallizes in space group C2221 with one polypeptide chain per asymmetric unit. The structure was determined to 2.08 Angstrom resolution [41]. The subunit displays the aββa sandwich fold characteristic of members of the nitrilase super family [41]. The majority of the secondary structure elements of HβAS-T299C are conserved in βAS of D. melanogaster which shares 64% amino acid sequence identity. In accordance with its oligomeric state in solution HβAS-T299C crystallized as a dimer that is assembled by application of crystallographic symmetry [41]. The missing interaction between the N-terminal helices reduces the dimer interface area by about one-fourth. Major contributions to the interface area are residues 185-192, 197-201, 213-222, 234-248, 263-278,322-324 and 350-384. Thirty hydrogen bonds and six salt bridges are formed at the interface, with 25 hydrogen bonds but none of the salt bridges are being conserved at the corresponding DmβAS interface. The active site of HβAS-T299C is located close to the monomer-monomer and (putative) dimer-dimer interfaces. The highly conserved cysteine, lysine and glutamate identified as catalytically crucial nitrilase-like enzymes (Cys233, Lys196 and Glu119) and other residues likely to be involved in catalysis or substrate binding are all derived from one subunit (Scheme 2) (Figure 6).
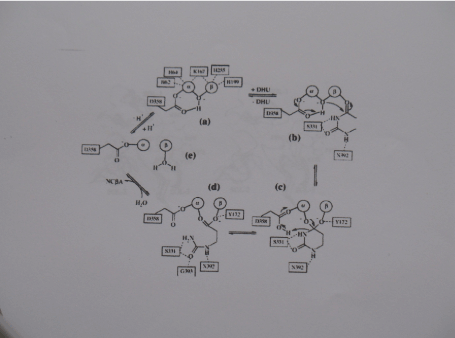
Scheme 2. Reaction mechanism of DHPase. Similar to the DHO-catalyzed reaction, hydrolysis of DHU is proposed via the Following steps: a-b) biding of DHU, b-c) nucleophile attack of its C4 atom by the zinc- Bridged hydroxyl and formation of thetetrahedral intermediate, C-d) breakage of the N3-C4 bond and formation of NCßA, d-c) product release and binding of a new solvent Molecule; e-a) the activation of a hydroxyl moiety. The given residue numbering Corresponds to DHPase from S. kluyveri. Only in (a) the complete zinc coordination is shown
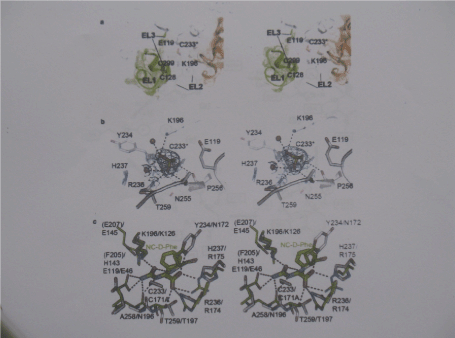
Figure 6. HßAS active site
- a) Stereo view of the active site entrance. The HßAS dimmer is shown in coloured white for subunit A, except for EL1 and the visible residues of EL2, shown in green, and gold for subunit B
- b) Close-up stereo view of the active site. © Stereo view of the superimposed active Site of HßAS (carbon atoms in gray) and of the Agrobacterium sp. N-carbamyl-D- Carbamyl amino acid amidohydolase double mutant C171A/V238A carbon atoms in green) in complex with its substrate N-carbamyl D-phenylalanine ( thicker sticks with carbon atoms in green)
Hydrolysis of βAS substrates is expected to follow the nechanism proposed for the closely related N-carbamoyl-D-amino acid amidohydrolase [42]. The role of the cysteine as catalytic nucleophile has been probed by site-directed mutagenesis for many nitrilase like enzymes [43]. Mutation of C233 to alanine renders the enzyme completely inactive. The substrate binding is expected to involve His237 and Arg238 as ligands of the carbonyl group and Lys196, Glu119,Ala258 and Tyr234 for interactions with the carbamoyl group.
Allosteric regulation and pH effect: Published data about properties of βAS from higher eukaryotic organisms
reveal diversity in their kinetic and allosteric behaviour, metal ion dependency and molecular size distribution. The rat enzyme (84% sequence identity with HβAS) appeared most complex, showing positive co-operativity towards the reaction substrates associated with assembly of more, larger oligomers, dissociation to smaller-sized oligomers (proposed trimers but probably rather representing a mixture of dimers and tetramers) and inactivation in response to the reaction product β-alanine but not βAIBA and competitive inhibition by many compounds of which some can and others cannot trigger association or dissociation [26,44]. On sentence is missing: However, the authors also noted that occasionally enzyme samples were obtained that were not responsive to allosteric effectors and showed an almost 20-fold increase in Km for NCßA [44] than the catalytic site , with substrate binding to this site promoting transition to the active enzyme conformation (T to R) (Figure 7).
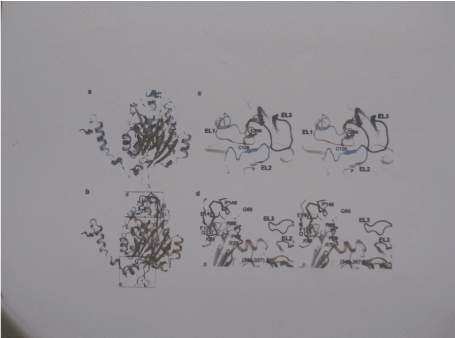
Figure 7. The subunit of human HßAS.
- Cartoon representation of the subunit fold with a-helices and ß-strand Coloured steel blue and coral, respectively. All secondary structure elements are Labelled.
- Superimposed subunits of HßAS ( coral) and ßAS from melanogaster. The latter Coloured gray except for the N-terminal 31 amino acids that are disordered in HßAS And the entrance loop EL1 (cyan), EL2 (Blue) and EL3 (dark blue)
- Close-up stereo view of the entrance loop region of the superimposed HßAS and ßAS From melanogaster subunits, coloured as in c)
- Close-up view of regions 61-70 and 345-357 that show larger structural deviations Between HßAS (subunits A and B of the dimmer shown as cartoon in coral and yellow, respectively) and ßAS from melanogaster (gray)
In contrast, calf liver βAS 84% sequence identity with HβAS did not show co-operativity towards NCβA or effector-responsive deviations from the homohexamer state [45], whereas the enzymes from Z. mays (60%), A, thaliana (59%) and D. melanogaster (64%) [31] exist in different states, but their responsiveness to reaction substrate or product , and co-operativity in their kinetic profile, were either not investigated or not observed, A major finding in HβAS [41] was the pH-dependence of its kinetic and oligomerization properties and adds to complexity of the allosteric regulation mechanism.
At pH 9.0, HβAS exists almost exclusively as homodimer, which is expected to contain catalytically impaired active sites due to lack of dimer-dimer interactions stabilizing the conformation of the active site entrance loops and resulting displacement of the highly conserved Glu207 from the active site.
At pH 7.4 , the enzyme retains nearly 50% of the optimal activity, observed at pH 6.5. Although the dimeric state is still predominant, a considerable proportion of HβAS exists already in a tetrameric or octameric state. NCβA binding shifts the equilibrium Further towards higher oligomers that contain an increasing number of catalytically competent active sites as at least the internal subunits are converted from T to R state.
Zinc binding site: Utilization of a di-metal centre for the activation of water molecule is quite common for enzymes hydrolysing carbon-nitrogen bonds. The di-metal centre is located on the surface of the cleft between the catalytic site and the dimerization domains and thus freely accessible to solvent molecules in the open state [31].With the exception of His226 all Zinc ligands are part of the loops decorating the C-terminal end of the β-sheet in the catalytic domain. The carboxyl group of Asp125 and a water molecule or hydroxyl ion serve as bridging ligands between the zinc which are separated by a distance of about 3.4 Angstrom.
Catalytic nechanism of β-alanine synthase: The cleavage of substrates N-carbamoyl-β-alanine and N-arbamyl-β aminoisobutyrate is achieved by activation of zinc-coordinated water molecule to the hydroxyl ion nucleophile, which subsequently attacks the bond between the carbon of the carbamoyl moiety and the nitrogen of the β-amino acid moiety of the substrate. The mutant H262A/E, H397N, E159D/A, R322A of the S. kluyveri enzyme have helped to elucidate the role of the mutated residues in catalysis [31].
Putative zinc binding site of HβAS: For rat βAS, binding of two zinc ions per subunit was shown by metal content analysis [44], and two putative zinc-binding sequences motifs were identified comprising His97 Glu101 and His158 as well as His280, His 293 and Glu297 [44]. Nevertheless, for both HβAS-T299C and DmβAS no zinc binding was observed in the crystal structures. Furthermore, from the putative zinc-ligating residues, Glu101 is exchanged to a lysine in The human enzyme, except Glu297, none are conserved in DmβAS. His97, His158 ang Glu101 cluster together on the protein surface, but are 6-14 Angstroms too distant From each other to allow Zn ion binding between them, and distances between His280, His293 and Glu297 are even larger. It is concluded that if rat, human and plant βAS indeed bind zinc ions, then it occurs at other than proposed sites [46].
βAS deficiency: βAS deficiency is an autosomal recessive disease caused by mutations in the βAS-encoding UPB1 gene. βAS-deficient patients exhibit elevated levels of NCβA and NCβAIBA, uracil, thymine and corresponding dihydropyrimidines in plasma, urine and cerebrospinal fluid [28]. They present with a variable clinical phenotype, from symptomatic To displaying severe neurological symptoms such as seizure, icrocephaly delayed myelination and developmental delay [46]. The symptoms are connected to β-alanine depletion and/or accumulation of NCβA.
Competing interest
The author declares that there are no competing interests associated with the manuscript.
References
- Schnackerz KD, Dobritzsch D, Lindqvist Y, Cook PF (2004) Dihydropyrimidine dehydrogenase: a flavoprotein with four iron-sulfur clusters. Biochim Biophys Acta 1701: 61-74. [Crossref]
- Wasternack C (1980) Degradation of pyrimidines and pyrimidine analogs--pathways and mutual influences. Pharmacol Ther 8: 629-651. [Crossref]
- Andersen G, Andersen B, Dobritzsch D, Schnackerz KD, Piskur J (2007) A gene duplication led to specialized gamma-aminobutyrate and beta-alanine aminotransferase in yeast. FEBS J 274: 1804-1817. [Crossref]
- Traut TW, Jones ME (1996) Uracil metabolism--UMP synthesis from orotic acid or uridine and conversion of uracil to beta-alanine: enzymes and cDNAs. Prog Nucleic Acid Res Mol Biol 53: 1-78. [Crossref]
- Bauer K, Salnikow J, de Vitry F, Tixier-Vidal A, Kleinkauf H (1979) Characterization and biosynthesis of omega-aminoacyl amino acids from rat brain and the C-6 glioma cell line. J Biol Chem 254: 6402-6407. [Crossref]
- Enna SJ, Gallagher JP (1983) Biochemical and electrophysiological characteristics of mammalian GABA receptors. Int Rev Neurobiol 24: 181-212. [Crossref]
- Wasternack C, Hause B (1987) 39 years of 5-fluorouracil. Pharmazie 42: 73-79. [Crossref]
- Heggie, Sommadossi JP, Cross DS, Huster WJ, Diasio RB (1987) Clinical pharmacokinetics of 5-fluorouracil and its metabolites in plasma, urine and bile. Cancer Res 47: 2203-2206. [Crossref]
- Toggenburger G, Felix D, Cuenod M, Henke H (1982) In vitro release of endogenous beta-alanine GABA, and glutamate, and electrophysiological effect of beta-alanine in pigeon optic tectum. J Neurochem 39: 176-183. [Crossref]
- Traut TW, Loechel S (1984) Pyrimidine metabolism: Individual characterization of the three sequential enzymes with a new assay. Biochemistry 23: 2533-2539. [Crossref]
- Kikugawa M, Kaneko M, Fujimoto-Sakata S, Maeda M, Kawasaki K (1994) Purification characterization and Inhibition of dihydropyrimideimase from rat liver. Eur J Biochem 219: 393-399. [Crossref]
- Kautz J, Schnackerz KD (1989) Purification and properties of 5,6-dihydropyrimidine amidohydrolase from calf liver. Eur J Biochem 181: 431-435. [Crossref]
- Jahnke K, Podschun B, Schnackerz KD, Kautz J, Cook PF (1993) Acid-base catalytic mechanism of dihydropyrimidinase from pH studies. Biochemistry 32: 5160-5166. [Crossref]
- Brooks KP, Jones EA, Kim BD, Sander BDEG (1983) Bovine liver dihydropyrimidine aminohydrolase: purification, properties and characterization as a zinc metalloenzyme. Arch Biochem Biophys 226: 469-483.
- Gojkovic Z, Jahnke K, Schnackerz KD, Piskur J (2000) PDY2 encodes 5,6-dihydropyrimidine aminohydrolase Which participates in a novel fungal catabolic pathway. J Mol Biol 295: 1073-1087.
- Gojković Z, Sandrini MPB, Piskur J (2001) Eukaryotic beta-alanine synthases are functionally related but have a high degree of structural diversity. Genetics 158: 999-1011. [Crossref]
- Walsh TA, Green SB, Larrinua M, Schmitzer PR (2001) Characterization of plant beta-ureipropionase and functional overexpression. Plant Physiol 125: 1001-1011.
- Gojkovic Z, Rislund L, Andersen B, Sandrini MPB, Cook, PF, et al. (2003) Dihydropyrimidine amidohydrolase and dihydroorotase share the sameorigin ans several enzymatic properties. Nucleic Acid Res 31: 1683-1692.
- Dudley KH, Butler TC, Bius DL (1974) The role of dihydropyrimidinase in the metabolism of some hydantoin and succinimide drugs. Drug Metab Dispos 2: 103-112. [Crossref]
- Kim GJ, Kim HS (1998) Identification of the structural similarity in the functional realated amidohydrolase acting on cyclic amide ring. Biochem J 330: 295-302. [Crossref]
- Syldatk C, May O, Altenvuchner J, Mattes R, Siemann M (1999) Microbial hydantoinases – industrial enzymes from the origin of life. Appl Microbiol Biotechn 51: 294-302.
- Waldmann G, Podschun B (1990) Assay for beta-ureidopropionase by high-performance liquid chromatography. Anal Biochem 188: 233-236. [Crossref]
- Sumi S, Imnaeda M, Kidouchi K, Ohba S, Hamjima N, et al. (1998) Population and family studies of dihydropyrimidinuria: prevalence, inheritance and risk of fluorouracil toxicity. Am J Med Genet 24: 336-340.
- van Kuilenburg ABP, Meinsma R, Zonenberg BA, Zoetkouw L, Baas F, et al. (2003) Dihydropyrimidinase deficiency and severe 5-fluorouracil toxicity. Clin Cancer Res 9: 4363-4367.
- van Kuilenburg ABP, van Lenthe H, van Gennip AH (1999) Radiochemical assay of beta-ureidopropionase using radio-labeled N-carbamyl-ß-alanine obtained via [2-14] 5,6 dihydrouracil. Anal Biochem 272: 250-253.
- Matthews MM, Traut RW (1987) Regulation of N-caebamyl-beta-alanine amidohydrolase, the terminal enzyme in pyrimidine catabolism, ligand-induced change in polymerisation. J Biol Chem 262: 7232-7237.
- Molenaar SH, Goehlich-Ratmann G, Engelke U, Humpfer E, Dvortsak O, et al. (2001) Ureido propionase deficiency; a novel inborn error of metabolism using NMR spectrossopy onurine. Magn Reson Med 6: 014-1017.
- Van Kuilenburg ABP, Meinsma R, Beke E, Assmann B, Ribes A, et al. (2004) Beta-ureidopropionase deficiency : a inborne error of pyrimidine degradation associated with neurological abnormalities. Human Mol Genet 13: 2793-2801. [Crossref]
- Lohkamp B, Andersen B, PiskurJ, Dobritzsch D ( 2006) The crystal structure of dihydropyrimidinases reaffirm the close relationship between cyclic amidohydrolases and explain their substrate specificity. J Biol Chem 281: 13765-13776. [Crossref]
- Pace HC, Brenner C (2001) The nirilase superfamily classification, structure and function. Genome Biol 2: 1-9.
- Lundgren S, Gojkovic, Piskur J, Dobritzsch D (2003) Yeast beta-alanine synthase shares structural scaffold and origin with di-zinc- exopeptidases. J Biol Chem 278: 51855-51862. [Crossref]
- Thoden JB, Phillips GN Jr, Neal TM, Raushel FM, Holden HM (2001) Molecular structure of dihydroorotase: a paradigm for catalysis through the use of a binuclear metal center. Biochemistry 40: 6989-6997. [Crossref]
- Seibert CM, Raushel FM (2006) Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 44: 6383-6391.
- Jabri E, Carr MB, Hausinger RP, Karplus PA (1995) The crystal structure of urease from Klebsiella aerogenes. Science 268: 998-1004. [Crossref]
- Abendroth J, Niefind K, May O, Siemann M, Sylsatk C, et al. (2002) The structure of L-hydantoinase from arthrobacter aurescens leads to an understanding of dihydropyrmidinase subsrate nd enantio specificity. Biochemistry 41: 8589-8597.
- Xu Z, Liu Y, Yang Y, Jiang W, Arnold E, et al. (2003) Crystal structure of hydantoinase from Burkholderia picketti at a resolution of 2.7 Angstrom; insights into the molecular basis ofenzyme thermostability. J Bacteriol 185: 4038-4049.
- Radha Kishan KV, Vohra RM, Ganesan K, Agrawal V, Sharma VM, et al. (2005) Molecular structure of D-hydantoinase from Bacillus sp. AR9: evidence for mercury inhibition. J Mol Biol 347: 95-105. [Crossref]
- Cheon YH, Park HS, Lee SC, Lee DE, Kim HS (2003) Structure-based mutational analysis of the active site residues of D- hydantoinase. J Mol B Enzym 26: 217-222.
- Lohkamp B, Andersen B, Piskur J, Dobritzsch D (2006) The crystal structures of dihydropyrimidinases reaffirm the close relationship between cyclic amidohydrolases and explain their substrate specificity. J Biol Chem 281: 13762-13776. [Crossref]
- Porter TN, Li Y, Raushel FM (2004) Mechanism of the dihydroorotase reaction. Biochemistry 43: 16285-16292. [Crossref]
- Maurer D, Lohkamp B, Krumper M, Widerstein M, Dobritzsch D (2018) Crystal structure of and pH dependent allosteric regulation of human β-ureidopropionase, an enzyme involved in anticancer drug metabolism. Biochem J 475: 2395-2418.
- Sakai T, Hasegawa T, Yamashita E, Yamamoto M, Kumasaka T, et al. (2000) Crystal structureof N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure 8: 729-737.
- Chen CY, Chiu WC, Liu JS, Hsu WH, Wang WC (2003) Structural basis for catalysis and substrate specificity of Agrobacterium radiobacter N-carbamoyl-D-amino acid amidohydrolase. J Biol Chem 278: 26194-26201. [Crossref]
- Matthews MM, Liao W, Kvalnes-Krick KL, Traut TW (1992) β-Alanine synthase: purification and allosteric properties. Arch Biochem Biophys 293: 254-263.
- Waldmann G, Cook PF, Schnackerz KD (2005) Purification and properties of beta-alanine synthase from calf liver. Protein Pept Lett 12: 69-73. [Crossref]
- Nakajima Y, Meijer J, Dobritzsch D, Ito T, Meinsma R, et al. (2014) Clinical, biochemical and Molecular analysis of 13 Japanese patients with ß-ureidopropionase deficiency demonstrates high prevalence of the c.977G>A (p.R3269) mutation. J Inherit Metab Dis 37: 801-812.









