Introduction: Since “nothing in biology makes sense except in the light of evolution” (T. Dobzhansky) and it develops through the interrelationships between living beings and their environment; the biological factors in the etiology of human infections can only be explained based on Darwin‟s evolutionary theory. Thus, environments as different as communities and hospitals originate diverse evolutionary processes in the causal agents of community and nosocomial infections; mainly in the development of resistance to antimicrobials.
Methods: To validate the possible significant qualitative and quantitative differences between the causal agents of nosocomial and community-acquired urinary tract infections, and their respective antimicrobial resistances; a clinical-epidemiological, descriptive, transversal, comparative and retrospective study was conducted; based on every urine culture and antibiogram performed during a one-year period in a Family Medicine Unit and a Regional General Hospital in Chihuahua, Mexico. For verification of the statistical significance of these differences, χ2, odds ratio, confidence interval and p-value were calculated with a 95 % reliability limits for all cases.
Results: Qualitative differences (genera, species and strains) were verified between the 27 causal agents of nosocomial and community urinary tract infections; with the most evolved bacteria and yeasts located in the hospital, having a 1.9 times higher resistance against almost every antibiotic, especially for Gram-negative bacteria and Escherichia coli. In all cases, these differences were statistically significant. For Gram-positive bacteria, less resistant in general, community strains had a 1.2 times higher resistance for two of four types of antimicrobials; but not significantly.
Conclusions: Regarding antimicrobial sensitivity, important differences were evidenced as for the Clinical Practice Guides recommendations; based on research from environments, societies and medical attention quite different from Mexico. Even if bacterial evolutionary processes and antibiotic resistance are global, they do not run in parallel nor are contemporary everywhere; and although similarities eventually prevail, they occur with local peculiarities; therefore, their consideration is decisive for a suitable and effective prevention and therapy of infectious diseases and their epidemic outbreaks.
Bacterial evolution, Antimicrobial resistance, Nosocomial and community- acquired urinary tract infections.
As stated by the World Health Organization (WHO): “Antibiotic resistance is a growing threat to public health care worldwide. Infections caused by antibiotic-resistance pathogens substantially increase the burden of both health care-associated infections and community- acquired infections” [1]. And within this context, “urinary tract infections (UTI) are one of the most common pathological conditions in both community and hospital settings: […] about 150 million people worldwide develop UTI each year […] Among the common uropathogens associated to UTIs development, UroPathogenic Escherichia coli (UPEC) is the primary cause” [2]. In Latin America, this germ is one of the most common pathogens with increasing resistance, by implicating a “progressive loss of antibiotic options, not only high-complexity acute care hospitals as well as the daily affects the community, as well” [3]. In Mexico, UTIs represent the second cause of infectious disease cases and have become a growing health problem, since the bacterium has developed increasing antimicrobial resistance [4]. “By changing the conditions for bacteria in hospitals (antibiotic stress) and environment (pollution), we are generating selective conditions which […] play a crucial role in the evolution of a broad spectrum of pathogenic or environmental bacteria” [5] and, as an essential part of this process, the development of multi- resistance to antimicrobials; increasing the incidence and mortality from infections, both nosocomial and those acquired in the community. For all this and according to T. Dobzhansky, since “nothing in biology make sense except in the light of evolution” [6], and that in cases like this “there is perhaps no better example of the Darwinian notions of selection and survival”; an evolutionary approach to the causative agents in the study of the UTIs can make sense regarding its virulence, pathogenesis, prevention and therapy.
Urinary tract infections (UTIs) involve the attack of a pathogenic germ, generally a bacterium, to any organ in the urinary tract (kidney, ureter, bladder, urethra), which causes an inflammatory process. Etiology is mostly bacterial, most commonly Gram-negative: Escherichia coli, Klebsiella psneumoniae, Proteus mirabillis, Pseudomonas aeruginosa; and less often Gram-positive, such as Staphylococcus saprophyticus. Especially in complicated UTIs, they can also be caused by opportunistic germs such as Candida spp and Mycobacterium tuberculosis [7].
The incidence of UTIs and the virulence, pathogenicity and response to antimicrobials of their causative agents vary considerably, depending on geographical regions, social and ecological environments, host characteristics (sex, age, sexual activity, education, and socioeconomic status), medical practices (pharmacological and surgical therapy, urinary catheter, etc.) and those acquired in the community or in the hospital. Such variations can only be explained satisfactorily thanks to evolution by natural selection of the causative microorganisms. In principle, based on Darwin's theory of evolution, who defined it synthetically as follows:
“As many more individuals of each species are born than can possibly survive; and as, consequently, there is a frequently recurring struggle for existence, it follows that any being, if it vary however slightly in any manner profitable to itself, under the complex and sometimes varying conditions of life, will have a better chance of surviving, and thus be naturally selected. From the strong principle of inheritance, any selected variety will tend to propagate its new and modified form [7]”.
While the development of multidrug-resistant bacteria is already a global threat to public health, as far as “very high rates of resistance were observed in both community-acquired and health-care-associated infections in all WHO regions”; it presents differentiated particularities between continents and countries, as well as within nations. For example, E. coli drug resistance to third-generation cephalosporins varies from 12% in Europe, 36 % in the United States, 68% in the Southeast Asia region, 70% in Africa, up to 77% in the Western Pacific región [1]. Latin America and the Caribbean ranks third in critical-priority with an average of 76.5% and a standard deviation of 8.1% of resistance to these antibiotics [3]. Therefore, “the global problem of antimicrobial resistance is particularly pressing in developing countries, where the infectious disease burden is high” [8,9] and “have experienced unfavorable trends in resistance” [10]; even greater than in the developed G20 nations (a group formed by the twenty major global economies) and the OECD (Organization for Economic Cooperation and Development) [11]. Even among USA, it has been detected that “resistance of Escherichia coli isolates to trimethoprim-sulfamethoxazole [TMP/SMX] varied significantly according to geographic region, ranging from high of 22 % in the western to low of 10 % in the northeast (p < 0.001)” [12]. Thus, they are particular “susceptibility patterns of uropathogenic Escherichia coli (UPEC) strains in their specific geographical locations or populations” [13].
The sex ratio for UTIs is 3:1 female-to-male. This difference begins from the first years of life. Both in women and minors under 18 years, the most frequent etiology is E. coli (> 80% of cases). Regarding its treatment, "drug-resistance is increasing for all antibiotics used in the treatment of UTIs" in women; leading to reinfection and relapses; and in children the use of fluoroquinolones is reserved [14-16].
Regarding medical practices, WHO has indicated that among the various factors that contribute to the emergence and global spread of resistance to antibiotics, stands out the “inappropriate antibiotic use and prescription in health-care settings and the community” 1; which includes the use of a narrow repertoire of antimicrobials and the misuse and abuse of it 9. Likewise, it has also been noted that “the global antibiotic use for UTI has led to resistant bacteria in both outpatient clinics and inpatient hospitals” [17]; although differentially between developed and developing countries.
“Escherichia coli it is the most frequent cause of urinary infection in the out-of-hospital environment” [18]; and also one of the common pathogens with increasing drug-resistance at community level for fluoroquinolones and third-generation cephalosporin [4]. For example, for TMP/SMX “resistance has increased to >20% in United States”, and 11.8% or even higher against fluoroquinolones; although it has not increased in 10 years for nitrofurantoin, continuing up to 92.2 %; and barely rose to 4 % against Amoxicillin-clavulanate [18]. Besides, “over the past 25 years, nosocomial transmission of commonly encountered, community-acquired, multidrug resistance organisms has been increasingly documented in developing countries” [8]; also with regional and international differences [17,19,20]. In the case of Mexico, “prevalence of resistance appears higher than those from developed countries, and even those observed in other Latin American Countries […] among the highest described in the literature”: 83.7 % to ampicillin, 63.2 % to carbenicillin, 55.5 – 60.6 % to the quinolone/fluoroquinolone family, 56.4 % to TMP/SMX [12,17].
On the other hand, as the WHO has pointed out, nosocomial infections constitute "the most frequent adverse event in health care", particularly because of "increased resistance of microorganisms to antimicrobials" [21]. And yet, in general, “UTIs are the most common hospital- acquired infections”, as in the case of Spain with “20.6 % of all nosocomial infections”, in which “the most frequently isolated microorganism was Escherichia coli (31.4%)” [22,23] and also from countries like USA [24] and other European nations [25]. But nevertheless, some studies do not differentiate between nosocomial and community-acquired UTIs [26], or do so inaccurately [27].
In hospitals the risk for UTI by catheterization is greater: over 80% of cases; and involves in addition a higher risk of being polymicrobial (28%) and 60 times more prone to bacteremia than not-catheterized patients [23]. And in its inside, their services also make the difference; particularly its Intensive Care Units, in which UTIs represent 20% to 50% of all infections; being E. coli the most commonly isolated microorganism with the highest resistance to β-lactams (96.7 %), maintaining a medium sensitivity to aminoglycosides (50.8 %) [18]. In the case of Mexico and specifically in the Hospital of this present investigation, being E. coli the main cause of all nosocomial infections (20%) up to 2014, fell to third place in 2018, with 15.8%; being surpassed by S. aureus and Acinetobacter baumannii [28]. Here, UTIs rank fourth among nosocomial infections with 12% of the total; caused by 24 bacterial species, but with a predominance of only three: E. coli (39.8%), Enterobacter cloacae (9.7%) and K. pneumoniae (6.8%). General multiresistance to antimicrobials of E. coli was 48.2 %; from 0 % to carbapenems and amikacin, intermediate to fluoroquinolones and sulfonamides (55%), and 100% against cefotaxime, ceftazidime and aztreonam [29].
Bacterial virulence is the quantitative aspect of its pathogenicity, measured by the number of microorganisms needed to cause disease. And the ability of pathogenic bacteria to cause disease in a susceptible host is determined by multiple virulence factors acting individually or together at different stages of infection [30]. “Bacteria, throughout evolution, have acquired […] factors or virulence determinants […] that favor their growth or survival during infection”; which activate in two phases: an early phase to promote colonization and invasion of its host (adherence, mobility and chemotaxis, invasion); and a late phase, properly pathogenic, to develop defense mechanisms (intracellular survival and mobility, evasion of the immune response and antigenic variation, submission and confrontation) [31].
In the case of bacteria, Richard Lenski, for example, has described their evolution process by natural selection [32]; and in particularly by a prolonged and refined experiment through 50,000 generations of 12 populations of E. coli [33]. He managed to identify three dialectically interrelated fundamental aspects in their evolutionary process: genetic variation; divergence of strains, species and populations; and fitness to the environment they colonize. All this through two main resources: coadapted genotypes and arms race (against antimicrobials, for example).
In general, “the bacterial mechanisms involved in pathogenicity and virulence have undergone a long evolutionary process dependent on the host-pathogen relationship. These changes are due to the selective pressure caused by the advent of antimicrobials in medicine. This pressure has forced microorganisms to adapt to changing conditions, acquiring or developing new mechanisms of pathogenicity and resistance […] When ignored, then the mechanisms by which we can fight bacteria remain unknown” [34].
A manifestation of the bacterial evolutionary processes, particularly important for medicine and public health, is resistance to antimicrobials, which become ineffective when the percentage of resistance of uropathogens exceeds 20-25% [35]. In the case of Spain, UPEC of patients from the community were almost 100 % sensitive to the introduction of ciprofloxacin and norfloxacin; but after four years of use, resulted in a progressive decrease in sensitivity down to 86 % [36]. In a study from 2002 to 2007, the resistance of this bacterium only decreased (although not significantly) for Nitrofurantoin (from 5.5 to 3.8 %); with “an overall increase in resistance” during this five-year period to other antibiotics; especially against β-lactams (56%) and fluoroquinolones (24.2%); with increases in a minor degree, from 1.7 to 3.9 %, against cefixime, tobramycin and clotrimazole; up to 9.6 and 13.7 % for ciprofloxacin and amoxicillin- clavulanic acid. In particular, UPEC developed during this period unacceptable values of resistance against clotrimazole, ciprofloxacin, norfloxacin and ampicillin [37].
Furthermore, a four-year (2003-2006) multicentric European study “showed a substantial increase in resistance to β-lactams, TMP/SMX, nitrofurantoin and quinolones […] and the rates of antibiotic resistance observed among nosocomial invasive isolates were higher” than those from the community [25]. And a comparative study from Tehran found that “overall resistance rates were higher among inpatients isolates compared to outpatients (p = 0.039) […] resistance to ampicillin, cefepime, and ceftazidime were significantly more prevalent in inpatients than outpatients (p = 0.037, 0.008 and 0.047, respectively)” [38].
About the bacterial evolutionary process, Lewontin, et al have warned its dialectic: “It is not the genes nor the environment that determine an organism, but a particular combination of both […] the organism depends on both genes and environment […] organisms are not only the product, but also the creators of their genotypes will evolve differently in the same Environment” [39], “a genotype does not give rise to a single type of development, but to a norm of reaction, a scheme of different types of development in different environments” [40].
In this context, the study of UTIs becomes a matter of utmost importance, by means of a comparison of the ones acquired in the community and the nosocomial ones; given the special and radically different environment or ecological niche that hospitals have developed, since on one hand, favorable factors are concentrated in there for the transmission, colonization and reproduction of pathogenic germs: the most serious infections, the most susceptible patients, a constantly controlled environment in terms of temperature, humidity, etc., effective modes of direct and crossed transmission, through contact between patients and health personnel, and above all, thanks to invasive procedures for diagnosis and treatment (punctures, venoclysis, catheters, probes, surgeries, etc.); and on the other hand and mainly, that the measures and the substances used profusely for cleaning, hygiene, asepsis and antisepsis and, even more, antimicrobials; all this contributes both to combating and decimating microorganisms, and, paradoxically, to the selection and development of empowered subpopulations against such resources.
In order to verify whether the qualitative and quantitative differences are significantly likely with respect to the causative agents of nosocomial urinary tract infections and those acquired in the community and, also, of their respective antimicrobial resistance; a clinical- epidemiological, descriptive, transversal, comparative and retrospective study was carried out; taking as community-acquired UTIs a sample (all of which positive cultures and antibiograms were achieved) of those diagnosed in outpatients of a Family Medicine Unit; and nosocomial UTIs as all those detected in patients admitted in a Regional General Hospital (both part of the Mexican Social Security Institute and located in the city of Chihuahua, Mexico) were included throughout the year 2018.
Urine cultures and susceptibility testing were performed by a team of Biological Chemists Parasitologists properly trained and with years of experience in managing a team VITEK® 2 Compact, an advanced-technology system for microbial identification and their sensitivity and resistance to antimicrobials, fully automated. Due to technical limitations, only the identification and classification of the causative agents of UTI could be performed; and no further typification, in particular, genomic and molecular studies that are essential to investigate bacterial evolutionary processes.
To check the statistical significance of the differences, χ2, odds ratio (OR), confidence interval and p-value were calculated, with a 95% reliability for all cases.
During 2018, in the Family Medical Unit (Unidad de Medicina Familiar, UMF) were diagnosed 3 797 UTIs (78.3% women and 21.7% men); of which 395 positive urine cultures (10.4%) with antibiogram were achieved, which constitutes the community sample. On the other hand, a total of 1 400 nosocomial infections were detected in the Regional General Hospital (HGR), with a rate of 10.2 cases per 100 hospital discharges. Of these, 185 (13.2%) were of urinary tract, ranking in the fourth place of importance (after pneumonia, those related to vascular lines and surgical site); in most cases, 58.9%, the causative agent could be identified by urine culture and its response profile to an antibiogram. An important risk factor, the bladder catheter, is much more frequent in the hospital: 84.3% of nosocomial UTIs were associated with catheterization. Thus, 20.5 community cases corresponded for each hospital case.
Overall, as shown in Table 1, 504 causal agents of UTIs belonging to 27 different species were identified in the UMF and the Hospital. In total, 395 of such were sampled from the community (78.4%) and 109 are nosocomial (21.6%), with a ratio of 3.6 by 1. The absolute majority of causing agents were bacteria (91.9%) and a minority were yeasts (8.1%). Among the former, Gram-negative bacteria were the most represented (82.1%), followed by Gram-positive (9.8%); differentiating in everything, qualitatively and quantitatively, according to its environmental location.
Table 1. Causal agents of Urinary Tract Infections by Medical Unit
Causal Agent |
Family
Medicine Unit |
Hospital |
Total |
|
# |
% |
# |
% |
# |
% |
Escherichia coli |
299 |
75.7 |
38 |
34.9 |
337 |
66.9 |
Klebsiella pneumonia |
21 |
5.4 |
11 |
10.1 |
32 |
6.3 |
Pseudomonas aeruginosa |
5 |
1.3 |
9 |
8.4 |
14 |
2.8 |
Acenitobacter baumannii |
0 |
0 |
3 |
2.7 |
3 |
0.6 |
Enterobacter cloacae |
0 |
0 |
2 |
1.8 |
2 |
0.4 |
Morganella morganii |
1 |
0.2 |
2 |
1.8 |
3 |
0.6 |
Proteus mirabilis |
8 |
2 |
1 |
0.9 |
9 |
1.8 |
Citrobacter freundi |
4 |
1 |
1 |
0.9 |
5 |
1 |
Enterobacter aerogenes |
4 |
1 |
0 |
0 |
4 |
0.8 |
Salmonella spp |
3 |
0.8 |
0 |
0 |
3 |
0.6 |
Serratia marcescens |
1 |
0.2 |
0 |
0 |
1 |
0.2 |
Shigella boundii |
1 |
0.2 |
0 |
0 |
1 |
0.2 |
GRAM NEGATIVE |
347 |
87.8 |
67 |
61.5 |
414 |
82.1 |
Staphylococcus epidermidis |
4 |
1 |
0 |
0 |
4 |
0.8 |
Staphylococus aureus |
2 |
0.6 |
4 |
3.7 |
6 |
1.2 |
Staphylococcus
haemolyticus |
1 |
0.2 |
2 |
1.8 |
3 |
0.6 |
Staphylococcus hominis |
1 |
0.2 |
1 |
0.9 |
2 |
0.4 |
Staphylococcus capitis |
0 |
0 |
1 |
0.9 |
1 |
0.2 |
Streptococcus agalactie |
8 |
2 |
1 |
0.9 |
9 |
1.8 |
Enterococcus faecalis |
7 |
1.9 |
12 |
11 |
19 |
3.8 |
Enterococcus faecium |
0 |
0 |
4 |
3.7 |
4 |
0.8 |
Corynebacterium ssp |
1 |
0.2 |
0 |
0 |
1 |
0.2 |
GRAM POSITIVE |
24 |
6.1 |
25 |
22.9 |
49 |
9.8 |
Candida spp |
21 |
5.3 |
1 |
0.9 |
22 |
4.4 |
Candida albicans |
3 |
0.8 |
7 |
6.5 |
10 |
2 |
Candida tropicalis |
- - - |
- - - |
6 |
5.5 |
6 |
1.2 |
Candida glabrata |
- - - |
- - - |
1 |
0.9 |
1 |
0.2 |
Candida krusei |
- - - |
- - - |
1 |
0.9 |
1 |
0.2 |
Candida guillermondii |
- - - |
- - - |
1 |
0.9 |
1 |
0.2 |
YEASTS |
24 |
6.1 |
17 |
15.6 |
41 |
8.1 |
Total |
395 |
100 |
109 |
100 |
504 |
100 |
In community-acquired UTIs, the predominance of bacteria is greater (93.9%), especially of Gram-negative (87.8%); with an equal percentage of Gram-positive and yeasts (all from the genus Candida): 6.1%. As for particular differences: while Escherichia coli caused three quarters of the UTIs in the community, in nosocomial infections it was only a little over a third; Klebsiella pneumoniae almost doubled its percentage in the community compared to hospital cases; and the nosocomial percentage of Pseudomonas aeruginosa was 6.4 times larger than in the community. However, in UTIs acquired in the community, neither Acinetobacter baumannii nor Enterobacter cloacae were detected. And on the other hand, four Gram-negative species present in the community were not detected in nosocomial UTIs: Enterobacter aerogenes, Salmonella spp, Serratia marcescens, or Shigella boundii. Using χ2, we tested the hypothesis of a statistically significant predominance of Gram-negative bacteria in the etiology of UTIs acquired in the community, with respect to the nosocomial (OR = 5.39, p = 0.000); and specifically of E. coli (OR = 5.9, p = 0.000).
On the contrary, both portions of hospital Gram-positive bacteria as well as yeasts were larger compared to those from the community: 3.7 and 2.7 times more, respectively. They included two absent species in UTIs from the community: Staphylococcus capitis and Enterococcus faecium. On the other hand, the Enterococcus causing nosocomial UTIs exceded 7.7 times the portion of the UTIs it caused in the community; and while S. epidermidis was not a cause of any nosocomial infection, hospital S. aureus exceeded 6.2 times the community strain. And in turn, the hospital percentage of Candida albicans was 8 times larger than in the community; however, from the Candida genus it was not possible to know more possible differences between their species, since the UMF did not specify most of them (87.5%). A much greater importance in UTIs acquired in hospital with respect to those from the community was proven to be statistically significant: for both Gram-positive bacteria (OR = 5.39, p = 0.000) and yeasts (OR = 2.86, p = 0.001).
For each of the 504 causative agents of UTIs identified in urine cultures, antibiograms were performed using 7,304 tests of 38 antimicrobials. Of the total 5,859 antibiograms of all bacteria from the community, 69.4 % were sensitive to antibiotics, 6.8 % with intermediate sensitivity and 23.8 % resistant to them. In turn, of the 1,349 hospital tests, 52.4% of the bacteria were sensitive, 2.9% with intermediate sensitivity and 44.7% resistant to antibiotics. Using χ2, the hypothesis of greater antimicrobial resistance of hospital bacteria was tested, with respect to the community (OR = 2.05, p = 0.000). However, with differences by bacterial Gram.
Among the 347 Gram-negative bacteria from the community, 69.4% were sensitive to antibiotics, 6.8 % with intermediate sensitivity and 23.7 % resistant to them. In the case of 67 Gram-negative bacteria from the hospital, the results were: 51 %, 2.8 % and 45.6 %, respectively. And such antimicrobial resistance of hospital Gram-negative bacteria was significantly higher, compared to those from the community (OR = 2.53, p = 0.000).
On the other hand, of the 24 Gram-positive bacteria in the community, 64.8 % were sensitive, 3.9 % with intermediate sensitivity and 31.3 % resistant to antibiotics. In turn, for the 25 hospital Gram-positive, these results were: 57.4 %, 3.5 % and 39.1 %, respectively. Also, in this case the resistance of Gram-positive bacteria from the hospital was significantly higher than those of the community (OR = 1.41, p = 0.057); although with smaller and contradictory differences than in the case of Gram-negative bacteria.
Regarding yeasts, of the total of 41 identified, all from the genus Candida, 92.2 % was sensitive to the four tested antifungals (100% to Amphotericin B and Voriconazole), 1.6 % with intermediate sensitivity, and 6.2 % presented resistance to two of them: Flucytosine (18.8 %) and Fluconazole (6.3 %). And although in this case the resistance was also higher in hospital yeasts over community ones; they were statistically contradictory: with failing χ2, OR = 3.98 and p = 0.08, perhaps due to an insufficient sampling.
By examining antimicrobial resistance according to its target in bacteria, in all four types considered it was superior in cases of nosocomial UTIs, compared to those acquired in the community; although with differences (Figure 1). In hospital cases, percentages of resistance were higher for antibiotics that inhibit the synthesis of folates (sulfonamides) with 65.3 %, and nucleic acid (fluoroquinolones) with 56.4 %; overcoming the respective of the community: 33.3% and 44.6 %.
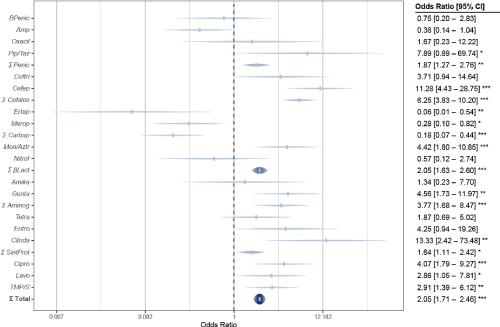
Figure 1. Total bacterial antimicrobial resistance in the Hospital vs Family Medicine Unit
And somewhat smaller differences in case for inhibitors of nucleic acid synthesis (fluoroquinolones) with 56.4% in hospital and 44.6% in the community; and inhibitors of protein synthesis (aminoglycosides, tetracyclines, macrolides, lincozamides, chloramphenicol, linezolides and streptogramins), with 33.2% hospital and 22% community. And there were also differences by bacterial Gram.
As shown in Figure 2, in Gram-negative bacteria differences in resistance were greater for antibiotics that inhibit cell wall synthesis: 47.4 % in the hospital and 14.6 % in the community (OR = 2.53, p = 0.000); and also for those that inhibit nucleic acid synthesis: 69.7 % hospital and 51.7 % community (with mean statistical significance: OR = 6.19, p = 0.000). The difference was somewhat smaller for folate synthesis inhibitors: 68.2 % from hospital and 56.3% from the community (OR = 3.43, p = 0.000). On the contrary, in terms of inhibition of protein synthesis, resistance was higher in the community (24.6 %) than in the hospital (23.6 %), but still not significant (OR = 1.14, p = 0.63).
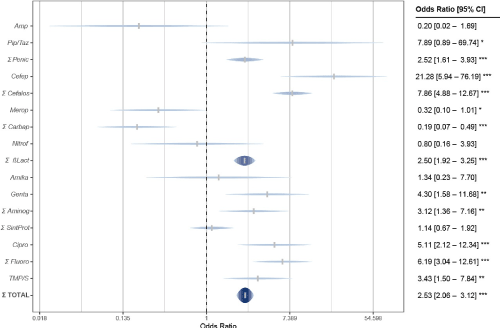
Figure 2. Antimicrobial resistance of Gram negative bacteria in the Hospital vs Family Medicine Unit
On the other hand, Gram-positive bacteria presented considerable differences and contradictions compared to Gram-negative in terms of their antimicrobial defense (Figure 3). The differences in resistance against nucleic acid synthesis inhibitors were larger: 44.6 % in hospital and 22.2 % in the community (OR = 2.09, p = 0.19); and to protein synthesis inhibitors: 44.3 % in the hospital and 25.7 % in the community (OR = 2.06, p = 0.023). On the contrary, for Gram-positive bacteria, although less resistant to the mentioned types of antimicrobials, community strains had a greater resistance against inhibitors of cell wall synthesis than the nosocomial strains: 34.6 % in the community and 26.5 % in the hospital (OR = 1.40, p = 0.25); as well against folate biosynthesis inhibitors: 41.7 % in the community and 33.3 % in the hospital (OR = 0.70, p = 0.25). However, despite the greater community resistance against cell wall and folate synthesis inhibitors, and perhaps due to the lack of statistical significance, in general Gram-positive bacteria from the hospital were more resistant to antibiotics (39.1 %) than those that caused UTIs in the community (31.3 %), and significantly: OR = 1.41 and p = 0.006.
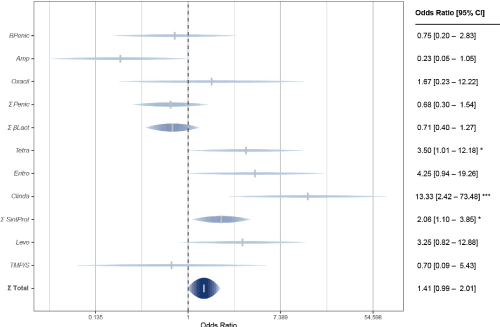
Figure 3. Antimicrobial resistance of Gram positive bacteria in the Hospital vs Family Medicine Unit
For the 27 species of microorganisms in the etiology of UTIs in both environments, the absolute predominance of Gram-negative bacteria (82.1 %) was mainly due to Escherichia coli with two thirds of the total (66.9 %); especially in the community (75.7 %), where it exceeded in more than double-fold its causality of UTIs to those from the hospital (34.9 %). Consequently, the patterns of bacterial sensitivity and resistance to antimicrobials were determined primarily by this causative agent.
The overall resistance to antimicrobials in hospital E. coli, 42.1 %, was much higher than the one achieved in the community: 23.5 %. Except when considering together carbapenems and antibiotics that inhibit protein synthesis, in all other cases the resistance rates were higher in hospital UTIs. However, such overall superiority of the resistance of E. coli strains in the hospital was confirmed significant, using χ2, OR = 2.39 and p = 0.000 (Figure 4).
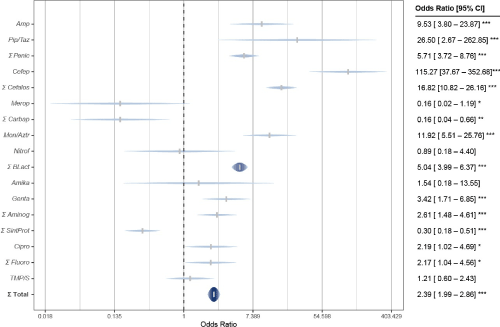
Figure 4. Escherichia coli antimicrobial resistance in the Hospital vs Family Medicine Unit
In particular, hospital E. coli resistance to penicillins of 50% exceeded 3.5 times to the one of strains from the community (14.1%); especially against Ampicillin: 81.6% compared to 31.7%, respectively. In the case of cephalosporins, their superiority was 6.4 times greater (69.3% versus 10.8%); especially for 68.4% in the hospital, given zero resistance against Cephalothin in the community; and 38 times more (68.4%) than the 1.8% community against Cefepime. With 68.4% in hospitals, resistance to monobactams (Aztreonam) was exceeded 5.3 times, compared to community strains (12.9%). In the case of Nitrofurantoin, the difference was smaller: 12.5 % versus 10.5 %. On the contrary, resistance to carbapenems (Imipenem and Meropenem) was 5.6 times greater in the community (14.5 %) than in the hospital (2.6 %).
However, resistance of E. coli hospital strains against inhibitors of cell wall synthesis (β- lactam and Nitrofurantoin) was 3.4 times greater (44.2 %) than the one achieved by community strains (12.8%); which was statistically significant: OR = 5.04 and p = 0.000.
In the case of protein synthesis inhibitors, E. coli strains from the community exceeded their resistance to them 2.3 times (39.8 %), compared to hospital strains (17.5 %); significantly: OR = 3.29 and p = 0.000. This is due, above all, to the zero resistance of hospital strains to Tigecycline and, nevertheless, their greater resistance to aminoglycosides: 26.3 % in the hospital and 12 % in the community, both for Amikacin (2.6 % and 1.7 %, respectively), as for Gentamicin (26.3 % and 12 %, respectively).
Regarding nucleic acid synthesis inhibitors (fluoroquinolones), resistance to them in hospital strains was higher (73.7 %) compared to those from the community (55.5 %); mainly in the case of Ciprofloxacin, with equal values and significantly: OR = 2.19 and p = 0.02. And in the case of folate biosynthesis inhibitors (sulfonamide), the minor difference between hospital resistance percentage of E. coli to Trimethoprim/Sulfamethoxazole (63.2 %) and the community (58.7 %) was not significant: OR = 1.21 and p = 0.60.
For its part, Klebsiella pneumoniae (which causes 6.3 % of the total UTIs) is resistant to Ampicillin at 100 % in both environments, reduced to 54.5 % with Sulbactam. Also 100% resistance to Amoxicillin, even with clavulanic acid. In the community, 27.8 % resistance to Piperacillin is reduced to zero with Tazobactam; which is resisted 18.2 % in the hospital. Thus, 57.6 % resistance to penicillins in hospital strains exceeded 35.9 % in community strains.
Against cephalosporins, hospital K. pneumoniae is 5.4 times more resistant (54.5 %), than in the community (10.1 %); mainly due to the zero resistance of the community strains against Cephalothin and Cefepime; and the considerable resistance to Ceftriaxone (54.5 %) from hospital strains. The same case for Nitrofurantoin: 0 % in the community and 45.4 % in the hospital. And against monobactams (Aztreonam), hospital strains resistance exceeded 3.6 times (54.5 %) compared to community strains (15 %). On contrast, zero resistance against carbapenems (Ertapenem and Meropenem) in hospital strains, contrasting with 15 % of community ones. Overall, the resistance of hospital K. pneumoniae of 50% to inhibitors of cell wall synthesis exceeded 2.4 times that of the community (20.6%).
Although in both environments resistance to Amikacin is zero, 22.7 % resistance to aminoglycosides of hospital strains exceeded 4.3 times the 5.3 % of the community; due to the very high resistance of the hospital against Gentamicin (45.5%). However, despite not being resistant to Tigecycline in the hospital, here 16.7% resistance to protein synthesis inhibitors slightly exceeded community strains: 13.7%.
As for antimicrobials that inhibit the synthesis of nucleic acid (fluoroquinolones), resistance to them present in hospital (63.6 %) quadrupled that from the community (15.8 %); especially against Ciprofloxacin. And in the case of folate biosynthesis inhibitors (sulfonamides), the resistance to Trimetroprim-Sulfamethoxazol in hospital, 54.5 %, was more than double that in the community: 25%.
Nevertheless, 40.7 % of global resistance to antimicrobials of hospital Klebsiella pneumoniae was more than double that of the community (18.9 %); due to their superiority against all, with the sole exception of the carbapenems. This was tested by χ2, resulting in OR = 2.96 and p = 0.000.
Although the incidence of UTIs by Pseudomonas aeruginosa is just 2.8 % of the total, its importance is remarkable for its very high antimicrobial resistance of hospital strains against all antimicrobials (75.7%) compared to the community (46.4%) and significantly (OR = 3.61, p = 0.000); mainly for achieving 100% resistance against Ampicillin (even with Sulbactam), Cephalothin, Ceftriaxone, Nitrofurantoin, Tigecycline and Trimethoprim-Sulfamethoxazole.
Gram-positive bacteria caused 9.8 % of the total UTIs, with almost two thirds of them (64 %) of the genus Enterococcus (fecalis and faecium) and almost one third (32 %) of the genus Staphylococcus (epidermidis, aureus, haemolyticus, capitis and agalactie). Overall, 39.1 % of hospital strains were resistant to antimicrobials and 31.3 % of community strains; however, such difference was of contradictory significance: with passing χ2, OR = 1.41, but p = 0.055. This is due to the fact that in two types of antimicrobials the resistance was higher in hospital and in two other it was lower than in the community.
Against inhibitors of cell wall synthesis (β-lactam and Nitrofurantoin), community strains achieved 34.6 % resistance and hospital strains 26.5 %; mainly due to the fact that in the community the genus Staphylococcus was 100 % resistant against: Penicillin G benzathine, Ampicillin, Amoxicillin (even with clavulanic acid), cephalosporins (Cefuroxime, Ceftriaxone and Cefepime) and Meropenem. Also, in the case of Trimetroprim-Sulfamethoxazole (folate biosynthesis inhibitor), which the community strains resisted in 41.7 % of the cases and 33.3 % in hospitals.
In contrast, Gram-positive bacteria in hospital showed greater resistance against protein synthesis inhibitors (44.3%), than in the community (25.7%); mainly against Clindamycin (83.3% vs. 23.5%), Erythromycin (73.9% vs. 43.7%) and Tetracycline (58.3% vs. 28.6%). As well with fluoroquinolones (inhibitors of nucleic acid synthesis): with 44.6% resistance in the hospital and 22.2% in the community; especially against Levofloxacin: 48% and 22.2%, respectively.
Since the hosts studied, both from the community and from the hospital, belong to the same population; the qualitative and quantitative differences observed in the pathogenesis of UTIs, and the virulence and response to antimicrobials by their causative agents, are mainly a result of competition between them and their interrelations with the environment. Once adapted to the latter, in order to survive, develop and reproduce; then microorganisms can spread from it, colonize and infect susceptible people; and within them, in their turn, pathogens must also adapt accordingly to the biological characteristics of the urinary tract. However, medical procedures, especially invasive devices and frequent use of antimicrobials, contribute decisively to this process. Especially in the hospital environment, it has been observed that all types of catheterized patients are at higher risk of contracting UTIs [18]; because the presence of the probe invariably induces pyuria [15]. Specifically, “the risk of acquiring bacteriuria is 3 % to 7 % per day of catheterization; thus patients with catheterization longer than 30 days have a prevalence of bacteriuria of 100%” [19].
In this regard, bacteria in general and Gram-negative in particular have been by far the most successful in the causation of UTIs; since they are better able to adapt, first, to the environment they come from and, from there, to the anatomical and physiological characteristics of the urinary tract by co-adapting their genotypes and developing defenses. That is why bacteria have limited yeasts to a minimum; especially in the community, because in the hospital, even with serious and immunocompromised patients, they act as opportunistic germs, increasing their virulence and pathogenicity.
In the community environment, competition is laxer and, therefore, relatively less proficient and less evolutionarily-developed germs (in their resistance to antimicrobials, in particular), such as E. aerogenes, Salmonella, Serratia or Shigella can be cause of UTI; and have not a chance in the hospital environment, much more aggressive and competitive. Conversely, bacteria in the evolutionary vanguard, such as A. baumanni and E. cloacae (multi-resistant to antimicrobials), well adapted to the hospital environment, did not appear in the community etiology. An intermediate situation is that of S. epidermidis, with cases of UTI in the community but not in the hospital, despite causing other types of nosocomial infections.
A determining factor for bacterial evolution has been the "arms race", thanks to the development of antimicrobial resistance; which depend on the class, modality, frequency and dosage these drugs are used in medical care; exerting a selective pressure among various species and strains. It has been proven in the case of Spain, for example, that “the increase in the consumption of antibiotics is recognized as one of the main causes of the increase in bacterial resistance to them”18. Conversely, reducing or discontinuing their use concomitantly decreases or nullifies such resistance; as is the case of penicillins and Nitrofurantoin which, with less hospital use, hospital bacteria showed less resistance to them, compared to community ones. This is due to the fact that antibacterial resistance is the result of structural and functional modifications characteristic of the causal or acquired agents and developed in response to the stress of the selective pressure by antimicrobials.
In this study a differential bacterial ability to resist antimicrobials was observed; which was globally superior in hospital species and strains, compared to those of community, with few exceptions. In particular, Gram-negatives are more capable to resist inhibitors of nucleic acid synthesis (fluoroquinolones) and those that inhibit folate biosynthesis (Trimetroprim- Sulfamethoxazole); yet are less so against inhibitors of cell wall synthesis (β-lactam and nitrofurantin); and have minimal resistance to antimicrobials that inhibit protein synthesis. Only in this case and of Carbapenems, resistance in the community was greater; however, without statistical significance. Particularly against Tetracycline and Meropenem, which is often used for outpatients.
On the other hand, the Gram-positive ability to resist antimicrobials is generally weaker than Gram-negative, except against protein synthesis inhibitors; especially against Clindamycin (mostly of hospital use), Erythromycin and Tetracycline (most used in the community). In this case, in general its maximum resistance is to those that inhibit the synthesis of nucleic acid and proteins; and higher against all antibiotics for hospital use.
There are several advantages that Gram-negative bacteria have compared to Gram- positive, in terms of antibiotic resistance. Regarding the mechanisms at an extracellular level, they are better able to produce biofilms; and for systems of quorum sensing (QS) and autoinducer chemical signaling between bacteria and communication between bacteria, operated by P. aeruginosa to induce resistance, or as Indol produced by E. coli to modify cellular functions related to gene expression, mobility, adhesion and pathogenicity, in response to environmental stress, including that caused by antibiotics. In the first instance, the cell wall or outer membrane exclusive to the Gram-negative, constitutes a more efficient selective barrier, by regulating the entry and expulsion of substances from the external environment; such as β- lactam, glycopeptides, ionophores antibiotics and others. Cell wall alterations inhibit the action of antimicrobials on this target; as is the case with the terminal end of peptidoglycan, which prevents the union of Vancomycin, or induces the degradation of β-lactam. Also restricts antibiotics that require porin channels in the membrane for access into the bacteria; such as β- lactam (cephalosporins and carbapenems) and fluoroquinolones and, in particular, are also protein structures highly resistant to detergents. E. coli has three main porins: PhoeE, OmpF and OmpC. For example, by stopping OmpC activity, in addition to preventing the entry of β-lactams into the bacteria, expulsion pumps are activated in the intermembrane space of Gram-negative bacteria (and to a lesser extent in the cytoplasmic membrane of Gram-positive); which prevent the required effective concentrations of antibiotics, detergents and other harmful substances. And to prevent its reflux into the interior, Gram-negatives operate a tripartite system with an internal membrane transporter, a periplasmic adapter protein and an external membrane channel. The one from enterobacteria such as E. coli is the most versatile. In contrast, mutant strains of P. aeruginosa, lacking an outlet pump, are more sensitive to gentamicin and ciprofloxacin. A main mechanism for antimicrobials inactivation is enzymatic hydrolysis, particularly against β-lactams (all penicillins, monobactams and cephalosporins); by penicillinases, carbapenemases, metallo-β- lactamases (Zn), cephalosporinases and oxacillinases. In particular, “the Enterobacteriaceae family associated to the nosocomial environment, has a high prevalence for this type of resistance mechanism”; although other negative and positive Gram can also activate it [41].
As for resistance mechanisms at the intracellular level, some bacteria use oxidation-reduction potential to evade antimicrobials. Enterobacteria and bacilli Gram-negatives, S. aureus and E. faecalis, can synthesize inactivating enzymes of the transfer group or transferases, which neutralize aminoglycosides, chloramphenicol, macrolides and others. Against antibiotics that inhibit protein synthesis, some bacteria evade their action thanks to ribosomal protection proteins (RPPs); a mechanism regulated by specific genes, more in Gram-negative: tet (M), (O), (S), (W), (Q), (T), (A) and (B); and fewer in Gram positives: tet (M) and (O). To nullify the effect of sulfonamides, S. aureus, for example, generates metabolic substances that compete with its active site in the bacteria; with a compound similar to PABA (para-amino benzoic acid), which is a precursor of bacterial folic acid. And like mechanisms of extracellular resistance, these are also subject to evolutionary processes by natural selection, thanks to genetic modifications [41].
In this sense, the predominance of E. coli in the etiology of UTIs is not by chance, but caused by multiple and diverse factors that confer it an evolutionary superiority when facing other germs which it competes against; like its extraordinary ecological plasticity, enabling it to quickly adapt to different environments, either as a free-living organism, a mutualist or a pathogenic commensal; and inside organisms, it can change and invade from one organ, apparatus or system to others. It was initially considered that the natural habitat of E. coli was restricted to the colon of mammals and birds, therefore transmitted by fecal contamination. However, existence of strains occupying other diverse niches has been verified, in particular acting as pathogens inhabiting other parts of the digestive tract, the urinary tract and blood. E. coli strains that can sicken humans are divided into two groups: diarrheagenic, which infect the intestinal tract; and those that cause extraintestinal infections, such as UTI, bacteremia, meningitis, surgical wound, etc. For example, the pathogenic strain O157: H7, very competitive, can successfully live in a large number of environments and hosts, overcoming environmental aggressions such as antiseptics and antibiotics; spreading epidemically in human populations, both within community and hospitals [42,43].
Thanks to the plasticity of the bacterial genome, they can develop resistance to environmental stress, caused by physical-chemical conditions, phages, detergents, antiseptics and antimicrobials; in order to survive it, adapt to the environment, reproduce and preserve the species. Thus, all antibiotic resistance mechanisms are genetically regulated, thanks to three fundamental processes: gene mutation, activation of specific regulatory genes and global transcription.
A classic example of protection and defense through gene mutations are the DNA gyrase and topoisomerase IV enzymes for DNA synthesis, which avoid damage by drugs such as quinolones; achieving resistance to fluoroquinolones with mutations to the DNA topoisomerases gyrA, gyrB and parC. It can also be achieved by regulating genetic transcription; being the main regulator of multi-resistance to disinfectants, organic solvents and antibiotics, the MarA operon or RamA gene, Sox Srob. And is decisive the ability to spread resistance genes through extrachromosomal elements, horizontally transferred between bacteria, even between different species; "which attaches importance to them in the evolution of bacteria by their easy horizontal transmission, generating resistance to different classes of antibiotics in the same extrachromosomal element"; such as plasmids ("conjugatives can be transferred intra or interspecies, a particularly important genetic phenomenon due to the selective pressure existing at the nosocomial level"), transposons or jumping genes, integrons ("granting selective advantages mainly to Gram-negatives") and genetic cassettes (“more than 130 have been recognized that encode resistance to virtually all families of antibiotics”) [41].
In the evolutionary process of resistance, gene cassettes have an important role, encoding both resistance to antiseptics and disinfectants (gene cassette qacE), as to antimicrobials (genes cassettes aac to aminoglycosides, cat to chloramphenicol, dfrA and B to trimethoprim, etc.) [30]. Also, genomic islands (GEIs), tools of bacterial horizontal transfer, play a crucial role in bacterial genome evolution and adaptation to changing conditions, in clinical, community or natural environments; they are metabolic and fitness islands, involved in the dissemination of variable genes, including antibiotic resistance and virulence and pathogenic genes leading to the generation of hospital “superbugs” as E. coli [44,45].
Different and changing processes facing different environments and different bacterial responses cause a periodic natural selection, in which a genotype selectively displaces others; when a favorable mutation arises by natural selection, not only that gene is replaced, but the complete genotype. In this way, UPEC strains 131 uropathogen-specific genes were found that are absent in fecal/commensal strains; 38 of which are involved in adaptation to growth within the host urinary tract. These genes, involved in environmental persistence of UPEC, are product of selective evolutionary pressures and patterns of gene transfer or loss [30]; for example: toxins (hlyD and cnf1), adhesins (papA and fimH, the most frequent virulence gene among UPEC [46]), iron transport (sitA) and specific antibiotics resistance genes [47]. This resistance is acquired mainly by horizontal transfer, even between bacteria of different species, by transformation, transduction and conjugation of genetic material [34]. And “UPEC strains with greater antibiotic resistance tended to express lower virulence traits” [38].
That occurs even within one species, for example, there is a dominant strain of E. coli per host, but it is only temporary: with the appearance of new genotypes, the best strain displaces the others. In the case of clonality, the "species" are constituted by a collection of independent evolutionary lineages, without having a common gene pool; and evolution will be given by substitutions of complete lineages, either by selection or by random genetic drift. In contrast, when high degrees of recombination occur, panmictic populations are obtained that approximate the populations and species of diploid organisms. Most bacteria are at an intermediate point [13]. Thus, it has been observed, for example, that temporal changes in the prevalence of community- acquired antimicrobial-resistant UTI by E. coli: “Of the 26 clonal groups first identified among period 3 isolates, 9 (35%) were no longer circulating during period 4. Of the 24 clonal groups infecting patients during period 4, three had not previously been identified” [48].
That is the reason why, in the case of UTIs, UPEC has become “a major global public health concern”, precisely because of its greater capacity for evolution and adaptation, developing the most varied and effective virulence factors related among themselves: (1) Morphological (biofilm, fimbria and pilicides, capsules, outer proteins of the membrane), there is a relationship between biofilm formation in UPEC from acute cystitis and recurrent UTI and a significant difference in antimicrobial susceptibilities of the biofilm and non-biofilm formers. (2) Motility and adhesion or chemotaxis (most cases of uncomplicated UTIs are community- acquired and gut colonization precedes access to urinary tract, by means of two opposite phenomena: adhesion and motility, for successful colonization and ascending infection), “the ability to adhere to host epithelial cells in the urinary tract represents the most important determinant of pathogenicity”. (3) To satisfy their nutritional (siderophores for ferrous iron uptake “is clearly required for successful infection”, in addition to zinc and manganese) and metabolic requirements (amino acids and peptides are the primary carbon sources for UPEC within the urinary tract, and peptide uptake is critical for fitness during infection; amino acid biosynthetic capacity is decisive to growth in urine). (4) Toxins (α-hemolysin, cytotoxic necrotizing factor-1, secreted auto-transporter toxin, protease involved in colonization). (5) Evasion of host defense mechanisms (specific antigens). (6) Genome (complete, annotated genome sequences are available for the different UPEC strains that cause asymptomatic bacteriuria, cystitis, pyelonephritis and sepsis; providing genome-wide insights into the presence of fitness/virulence factors) and gene expression (genomic sequence and pathogenicity islands, such as HGT, one of the fastest means of dissemination of fitness or virulence factors among bacteria; global gene expression regulators, such as the virulence factor PhoP that controls the expression of around 11% of the genome and affects acid resistance by modulating polarization state inner membrane; histone-like nucleoid structuring protein; sigma E that facilitates transcription of genes required to combat envelope stress; host factor Q-mediated riboregulation and Quorum-sensing system QseBC are critical for coordinate gene expression during UPEC infection). [2,34,38,47,49].
In Spain, for example, the etiology of community-acquired UTIs by E. coli was 92%, 2.3 times higher than the 39% nosocomial rate [23]. Here, “the sensitivity of out-hospital E. coli was higher than hospital strains, with higher differences on cephalosporins, nitrofurantoin and against ampicillin-sulbactam and amoxicillin-clavulanate”; there were “in general minor differences in the most commonly used antimicrobials in the community, such as aminopenicillins, cotrimoxazole and quinolones”; however, eventually “originally observed sensitivity differences decreased between hospital and community-acquired strains” [31]. In the case of the USA, comparing their resistance to antibiotics, the hospital E. coli was superior to those acquired in the community for third and fourth generation cephalosporins, and lower against fluoroquinolones and carbapenems [26]. Against ciprofloxacin, gentamicin andTMP/SMX, in Colombia “resistance was more frequent in outpatients (emergency and external consultation) than in hospitalized patients” [50]. Penicillins have lower activity against E. coli, due to β-lactamases; in particular, the prevalence of ESBL (extended-spectrum β-lactamases)-producing strains is superior in nosocomial infections [23]. In fact, “hospitalization is a major cause of the development of infection by ESBL-producing bacteria” [20]. However, “the increasing frequency of ESBL phenotypes in the community is an emerging problem” [18,19].
In Mexico the uropathogenic E. coli (UPEC) strains belong to a limited number of O serogroups: O1, O2, O6, O18 and O75; being the most frequent in UTIs the O25 ETEC. Likewise, E. coli strains have been distinguished by their antimicrobial susceptibility profile in four groups: A (27.8%), B1 (8.4%), B2 (36%) and D (27.8%). They are commensal A and B1; while most strains causing extraintestinal infections, especially UTIs, are groups B2 and D to a lesser extent. Most multidrug resistance strains were from group B2 (52.7%). And “recently, an E. coli clone, O25-ST131, has emerged worldwide as an important cause of community-onset UTI, with high virulence potential produces ESBL” [43].
According to an Iranian study, the resistance of UPEC to TMP/SMX, cefotaxime and amoxicillin-clavulanic acid is related to the virulence genes fimH (the most frequent among UPEC), papa, hlyD , cnf-1, sitA and tsh; whose diverse possession among their phylogenetic groups gives different degrees of resistance to antimicrobials, being higher in Type A (up to 62.4%) and lower in Type D (up to 14.1%) [46]. And “the emergence and dissemination of antibiotic-resistant E. coli might be the result not only of the selection of different mutant strains generated by local antimicrobial prescription habits, but alternatively or concomitantly by clonal spread” [25].
In the case of Mexico, it has been shown that E. coli must acquire virulence determinants that enable it to cause UTIs; thus, UPEC strains are genetically more diverse than diarrheagenic E. coli. Seventy-two patterns of virulence markers were distributed across eight E. coli phylogenetic groups, with four predominant serogroups, particularly O25 [51]. Regarding multi- resistant clone O25-ST131, producer of the ESBL enzyme CTX-M-15 and causative of meaningful morbi-mortality by UTI, “molecular epidemiological studies suggested that the sudden worldwide increase of CTX-M-15 producing E. coli was mainly due to a single clone (ST131) […] spread across different continents” [52]. Genes of ESBLs found with higher frequency among UPEC were: blaTEM, blaSHV, and blaCTX-M group 1, CIT (plasmid-mediated AmpC β-lactamase), and blaOXA-like [51]. Since these particular genes and specific mutations indicate a high bacterial potential to multidrug resistance; E. coli fourteen genes were also associated with resistance to aminoglycosides, sulfonamides and quinolones, blaTEM-1 and sul2 are present in almost all strains [53].
As already noted, the study in general of the evolution by natural selection of the causative agents of infections, both in the community and in hospitals, and in particular, “the molecular and structural study of their antibiotic resistance, will allow us to recognize the risk points in our infection control policies and attain a more effective prevention against the production and dissemination of resistance” [41]. Specifically, it makes possible to track the variations in the microbiota and its ever-changing response to antimicrobials; with the purpose of adapting and streamlining antibiotic therapy and, with it, preventing the emergence of outbreaks and, if they occur, to know the best weapons to combat them. And as for medical care, it provides an accurate knowledge of microbial sensitivity in a given time to make it more effective.
According to the results of the present study, in the First Level of medical care of UTIs in the community, the generality of their causative agents presented the greatest sensitivity to: Amikacin (96.7%), Piperacillin-Tazobactam (93.4%) and Carbapenems (70.8%); corresponding in particular to Gram-negative bacteria and, especially, Cefepime (93.5%). And in the case of Gram-positive: Glycopeptides, such as Vancomycin and Teicoplanin (100%), Amikacin (96.7%), Linezolid (93.7%) and Piperacillin - Tazobactam (93.4%).
As for nosocomial UTIs, their causal agents in general presented the greatest sensitivities to: Carbapenems (94.1%), Amikacin (93.5%) and Tigecycline (85.2%); corresponding mostly to Gram-negatives, in addition to Piperacillin-Tazobactam (78.9%) and, to a lesser extent, Tigecycline (79.7%). And in the case of Gram positive: Linezolid (100%), Glycopeptides (91.7%) and Clindamycin (83.3%). With this, important differences are noticed regarding the antibiotic therapy recommendations contained in the current Clinical Practice Guidelines. For example, the University of Michigan Health System – GPC Sector Salud Mexico, “recommended as first-line treatment Trimethoprim-sulfamethoxazole, TMP/SMZ (160/800 twice daily for 3 days), and as an alternative... Nitrofurantoin for 7 days (100 mg twice daily)”. And for second-choice treatment… ciprofluoxacin 250 mg every 12 hours for 3 days”14. And in particular, “in children three months or older… they are first-choice antimicrobials: Trimetroprim-sulfamethoxazole, amoxicillin, amoxicillin/clavulanic acid, nitrofurantoin, or first or second generation cephalosporins ” [16].
However, our study in the case of the city of Chihuahua, Mexico, has revealed even very high resistance against such antimicrobials, except in specific cases. This evidences bacterial evolutionary processes and their antibiotic resistance that, even if they are generally global, do not take place in parallel nor are they contemporary everywhere. Therefore, the differences with respect to the recommendations of the Clinical Practice Guidelines are due to the fact that they are based on investigations of societies, countries, environments and ambulatory and hospital health care very different from those in Mexico; and although their similarities end up imposing themselves over time, the bacterial evolutionary processes and the development of antimicrobial resistance take place with local particularities that, nevertheless, their consideration is decisive for an adequate and effective prevention and therapy of infectious diseases and of its epidemic outbreaks.
- WHO (2017) Prioritization of pathogenesis to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. Geneve, World Health Organization. Switzerland
- Maria E Terlizzi, Giorgio Gribaudo, Massimo E Maffei (2017) UroPathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front Microbiol 8: 1566.
- Health World Organization (2018) Panamerican Health Organization: Recommendations for implementing antimicrobial stewardship programs in Latin America and the Caribbean. Washington, DC.
- General Health Council (2014) Diagnosis and treatment of uncomplicated acute pyelonephritis in adults. Master Catalog of Clinical Practice Guidelines: SS-185-10.
- Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, et al. (2009) Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33: 376-393.
- Stephen J Gould and Richard C. Lewontin (2017) Biological adaptation. Scientific World 3: 214-222.
- MAPPA GUIDES (2010) Antibacterial Management of Infectious Processes in the Adult Patient: Diagnosis and antibacterial treatment of urinary tract infections. National Academy of Medicine of Mexico. Mexico, DF.
- Charles Darwin MA (1861) The origin of species by means of natural selection, or the preservation of favored roots in the struggle for life. D. Appleton and Company, New York.
- Iruka N Okeke, Ramanan Laxminarayan, Zulfigar A Bhutta (2005) Antimicrobial resistance in developing countries. Part I: recent trends and current status. The lancet 8: 481-493.
- Iruka N Okeke, Keith P Klugman, Zulfigar A Bhutta (2005) Antimicrobial resistance in developing countries. Part II: strategies for containment. Infection. The lancet 5: 568-580.
- OECD (2018) Stemming the superbug tide.
- Kalpana Gupta, Daniel F Sahm, David Mayfield, Walter E Stamm (2001) Antimicrobial resistance among uropathogens that cause community-acquired urinary tract infections in women: a nationwide analysis. Clin Infect Dis 33: 88-94.
- Molina-López J, Aparicio-Ozores G, Ribas-Aparicio RM, Gavilanes-Parra S, Chávez-Berrocal ME, et al. (2011) Drug resistance, serotypes and phylogenetic groups among uropathogenic Escherichia coli including O25-ST 131 in Mexico City. J Infect Dev Ctries 5: 840-849.
- General Health Council (2009) Diagnosis and treatment of acute, uncomplicated infection of the urinary tract in women. Clinical Practice Guide IMSS-077-08. Mexico.
- General Health Council (2016) Prevention, diagnosis and treatment of low urinary tract infection during pregnancy in the First Level of care. Master Catalog of Clinical Practice Guidelines: IMSS-078-08. Mexico City.
- General Health Council (2008) Prevention, diagnosis and treatment of uncomplicated urinary tract infection in children under 18 years of age. Master Catalog of Clinical Practice Guidelines: SS-027-08. Mexico City.
- Soraya S Andrade, Helio S Sader, Ronald N, Jones (2006) Increased resistance to first-line agents among bacterial pathogens isolated from urinary tract infections in Latin America: time for local guidelines? Mem Inst Oswaldo Cruz 101: 741-748.
- Thomas A Waller, Sally Ann L Pantin, Ashley L, Yenior Urinary tract infection antibiotic resistance in the United States. Prim Care Clin Office Pract 3: 455-466.
- Jorge Alberto Cortez, Diego Perdomo (2015) Gu ed Clinical Practice Guidelines on diagnosis and treatment of infection in women uncomplicated community - acquired urinary tract. Acceso al artículo o revista 63: 565.
- Colodner R, Rock W, Chazan B (2004) Risk factors for development of extended-spectrum β-lactamase-producing bacteria in nonhospitalized patients. Eur J Clin Microbiol Infect Dis 23: 163-167.
- https://www.who.int/gpsc/country_work/burden_hcai/en/
- Savas L, Guvel S, Onlen Y, Savas N, Duran N (2006) Nosocomial urinary tract infections: micro-organisms, antibiotic sensitivities and risk factors. West Indian Med J 55: 188-93.
- Pigrau C (2013) Nocosomial urinary tract infections. Enferm Infecc Microbiol Clin 31: 614-624.
- Sanjay Saint, Christine P Kowalski, Samuel R Kaufman (2008) Preventing hospital-acquired urinary tract infection in the United States: a national study. CID 46: 243–250.
- Gian Carlo Schito, Kurt G Naber, Henry Botto (2009) The ARESC study: an international survey on the antimicrobial resistance of pathogens involved in uncomplicated urinary tract infections. Int J Antimicrob Agents 34: 407-413.
- John E delsberg, Derek Weycker, Rich Barron (2014) Prevalence of antibiotic resistance in US hospitals. Diagn Microbiol Infect Dis 78: 255-262.
- Brian Montenegro-Diaz, Rosita Tafur-Ramirez, Cristian Diaz-Velez, Jorge Fernandez- Mogollon (2016) Intrahospital infections of the urinary tract in critical services of a public hospital in Chiclayo, Peru (2009-2014). Acta Med Peru 33: 189-194.
- Hector Salazar-Holguin (2018) Acinetobacter baumannii antibiotic multiresistance evolution in hospital acquire infections, c linical data from a six-year study in Mexico. American Journal of Clinical Microbiology and Antimicrobials. 1: 1-7.
- Hector Daniel Salazar-HolguÃn, MarÃa Elena Cisneros-Robledo: Resistance to antimicrobials of causative agents of the main nosocomial infections. Rev Med Inst Mex Seguro Soc 54: 190-195.
- Hsing-Ju Wu, Andrew HJ, Wang, Jennings MP (2008) Discovery virulence factors of pathogenic bacteria. Curr Opin Chem Biol 12: 93-101.
- Sonia Junquera, Elena Loza, Fernando Baquero (2005) Evolution of the sensitivity pattern of Escherichia coli isolates in urine cultures from the hospital and extrahospital environment. Enferm Infecc Microbiol Clin 23: 197-201.
- Richard E Lenski (1992) Evolution, experimental. Encyclopedia of Microbiology. Academic Press, Inc. USA.
- Richard E Lenski (2011) Evolution in action: at 50,000 - Generation salute to Charles Darwin. Microbe 6:1.
- Mara Elena Cárdenas-Perea, Othón Rafael Cruz López, José Luis Gándara-RamÃrez, Marco Antonio Pérez-Hernández (2014) Factors of bacterial virulence: the 'intelligence' of bacteria. Elements 94: 35-43.
- Gupta K, Scholes D, Stamm WE (1999) Increasing prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in women. JAMA 281: 736.
- Sauca G, Gallés C, Gasós MA (1997) Changes in the sensitivity of Escherichia coli to 6 antimicrobial agents during the last 12 years. Group of Microbiologists of County Hospitals of Catalonia. Aten Primaria 19: 226-229. [Crossref]
- Jose Sanchez Merino, Cristina Guillan Maquieira, Carlos Fuster Foz (2008) Evolution of resistance to antibiotics of Escherichia coli in urine samples from the community. Arch Esp Urol 61: 776-780.
- Mohsen Tabasi, Mohammad Reza Asadi Karam, Mehri Habibi (2015) Phenotypic assays to d and end virulence factors of uropathogenic Escherichia coli (UPEC) isolates and Their correlation With antibiotic resistance. Osang Public Health Res Perspect 6: 261-268.
- RC Lewontin, Rose S, Kamin LJ (2009) It's not in the genes. Racism, genetics and ideology. Drakontos Pocket. Barcelona.
- Richard C Lewontin (2000) Genes, organism and environment. The cause and effect relationships in biology. GEDISA Editorial. Barcelona.
- Claudia Troncoso, MÃnica Pavez, Andras Santos (2017) Structural and physiological implications of bacterial cell in antibiotic resistance mechanisms. Int J Morphol 35: 1214-1223.
- Valeria Souza, Amanda Castillo, Martha Rocha (2001) Evolutionary ecology of Escherichia coli. INCI 26: 10.
- José Molina López, Ángel Manjarrez Hernández (2015) Urinary tract infections: Escherichia coli. Faculty of Medicine, UNAM, 2015.
- Hall RM, Collis CM (1998) Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist Updat 1: 109-119. [Crossref]
- Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, et al. (2009) Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33: 376-393. [Crossref]
- Abdollah Derakhshandeh, Roya Firouzi, Mohammad Motamedifar (2015) Virulence characteristics and antibiotic resistance pattern s among various phylogenetic groups of uropathogenic Escherichia coli isolates. Jpn J Infect Dis 68: 428-431.
- Josa Molina Lapez, Ãngel Manjarrez Hernandez (2015) Urinary tract infections - Escherichia coli. Department of Public Health, Faculty of Medicine, UNAM.
- Sherry P Smith, Amee R Manges, Lee W Riley: Temporal changes in the prevalence of community-acquired antimicrobial- resistat urinary tract infection affected by Escherichia coli clonal group composition. Clin Infect Dis 46: 689-695.
- Subashchandrabose S, Mobley HLT (2015) Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol Spectr 3: 4.
- Norton Perez Standard Pavas, Emma Isabel Rodriguez (2011) Antibiotic resistance in Escherichia coli with β- lactamases of expectro extended in a hospital Colombian Orinoco Infectio 15: 147-154.
- Gloria LUZ Paniagua-Contreras, Eric Monroy-Perez, Areli Bautista (2018) Multiple antibiotic resistances and virulence markers of uropathogenic Escherichia coli desde Mexico. Pathog Glob Health 112: 415-420.
- Peirano G, Pitout JD (2010) Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents 35: 316-321.Agents 35: 316-321. [Crossref]
- Wioletta Adamus-Bialek, Anna Baraniak, Monika Wawszczak (2018) The genetic background of antibiotic resistance among clinical uropathogenic Escherichia coli strains. Molecular Biology Reports 45: 1055-1065.




