Abstract
Background: TP53 gene is a tumour suppressor gene located in the short arm of chromosome 17. TP53 plays a pivotal role in maintaining genomic stability in response to DNA damage. It is mutated in more than 50% of the human cancer. TP53 mutation (TP53mut) is commonly associated with therapy-related acute myeloid leukemia (AML) and complex karyotype. The incidence of TP53mutn was between 5-10% in de novo AML. This study described the clinicopathologic features of TP53mut AML and their clinical outcome in an Asian cohort.
Method: 166 consecutively cell-banked marrow samples of AML were tested for TP53 mutations using a next generation sequencing platform. Baseline disease characteristic and clinical outcomes were retrospectively collected with approval granted by institutional review board.
Results: 9/166 had TP53mut (5.4%). 6/9 of TP53mut AML was associated with complex karyotype (p<0.001). The remaining 3 cases were two acute promyelocytic leukemia (APML) and one with normal cytogenetics. TP53mut was mutually exclusive with FLT3 mutation. Two cases had concomitant NPM1 mutation and another two had ASXL1 mutation. TP53mut AML appeared to be associated with TET2 mutation (3/9, 33.3%) compared to 10.2% of the TP53wt (p=0.069). TP53mut AML had a lower CR rate compared to TP53wt (28.6% vs 84.3%, p=0.003). Only two cases achieved CR, one was an APML who remained in continuous remission after 4 years. The other case achieved CR after allogeneic transplant but, relapsed 8 months later. TP53mut AML was significantly associated with inferior OS compared to TP53wt (p=0.001). 0.8 months (95%CI: 0.159 - 1.484) and 36.8 months (95%CI: 0.000 - 92.676) respectively. It remains an independent predictor of OS in multivariate analysis that include cytogenetic risk, WBC, LDH and BM blasts (HR 43.07, 95%CI: 6.87-272.41, p<0.001).
Conclusion: Our results confirmed the extremely dismal prognosis of TP53mut AML. TP53 mutation testing should be included as part of the pre-chemotherapy workout since this is not a standard practice as yet. The significance of its association with TET2 mutation requires further exploration in a larger study cohort.
Introduction
Acute myeloid leukaemia (AML) is a clonal disorder, characterized by accumulation of immature myeloid progenitor cells [1,2], resulting from uncontrolled cell division and impaired differentiation. It is the most common form of acute leukemia affecting adults [2]. AML is a heterogenous group of condition, consisting chromosomal abnormalities, recurrently mutated genes and microRNA deregulations [3], and it has varied clinical outcome. Risk stratification is conventionally done based on cytogenetic profile and it divides patients into three categories: favourable, intermediate and poor [4]. However, a significant number of AML patients with normal karyotype are classified as intermediate risk despite differing clinical outcomes due to underlying molecular mutations [5]. Hence, more recently, recurrent genetic mutations have been incorporated into risk stratification to refine individual prognosis and management of AML [2].
With next-generation sequencing (NGS), the discovery of recurrent molecular mutations found in AML patients with normal cytogenetics is accelerated [1]. An overview of these recurrent mutations is now available due to the presence of NGS. Such parallel sequencing of a large number of genes reduces cost and time needed [1,6], making molecular genetic testing a viable part of a potential diagnostic measure. After much study on these recurrent gene mutations and their prognostic significance, NPM1, CEPBA and FLT3-ITD have been incorporated into the 2010 recommendations by European LeukemiaNet (ELN) [7].
In the recent 2017 ELN guidelines, TP53, the most commonly mutated gene in human tumours [8,9], has also been included. TP53 is a tumour suppressor gene located in the short arm of chromosome 17 and it plays a pivotal role in maintaining genomic stability in response to DNA damage [10]. It activates DNA-repair programs and triggers cell-cycle arrest. Some of the common forms of inactivation of TP53 include deletion of the short arm of chromosome 17 and missense point mutations. The incidence of TP53 mutation has been reported to be between 5-10% in de novo AML [10]. It is also associated with therapy-related AML, complex cytogenetics, low blast counts, and an under-representation of concomitant mutations in FLT3, RAS, NPM1, and RUNX1 [10]. These patients exhibit lower response rates to intensive chemotherapy and shorter complete remission (CR) durations [11,12], translating into inferior survival in both younger and older patients [13].
In this study, we aim to evaluate the prognostic significance of TP53 mutational status in a South East Asian cohort. Singapore is a racially diverse country with the majority of its citizens being from around the Southeast Asian region, providing unique population demographics for this study.
Material and methods
Study population
After obtaining approval from the institutional review board, pre-treatment diagnostic bone marrow (BM) samples of 166 AML patients were obtained from the archives of the Department of Haematology-Oncology, National University Hospital, Singapore. These patients were diagnosedwith AML between 2001 and 2016, in accordance with the 2001, 2008 and 2016 WHO classifications. The demographic, laboratory and clinical data were recorded retrospectively.
Next-generation sequencing mutational analysis
Genomic DNA was extracted from the consecutively cell-banked marrow samples of AML. In brief,targeted DNA sequencing was performed using the TruSeqCustom Amplicon assay for the TruSight Myeloid SequencingPanel (Illumina) as previously described [14]. The 54 genes assessed via the TruSight Myeloid Sequencing Panel includeABL1, ASXL1, ATRX, BCOR, BCORL1, BRAF, CALR, CBL,CBLB, CBLC, CDKN2A, CEBPA, CSF3R, CUX1, DNMT3A, ETV6, EZH2, FBXW7, FLT3, GATA1,GATA2, GNAS, HRAS,IDH1, IDH2, IKZF1, JAK2, JAK3, KDM6A, KIT, KMT2A,KRAS, MPL, MYD88, NOTCH1, NPM1, NRAS, PDGFRA, PHF6, PTEN, PTPN11, RAD21, RUNX1, SETBP1, SF3B1,SMC1A, SMC3, SRSF2, STAG2, TET2, TP53, U2AF1, WT1and ZRSR2. The TruSeq Amplicon (BaseSpace workflow, V.1.1.0.0) was used to generate the BAM and VCF files.Subsequently, the VCF files were annotated using IlluminaVariantStudio (V.2.2). Only variants that met the following criteria were included: non-synonymous; only variants insidegenes; quality >99 and variant allele frequency ≥ 10%.Variant-calling was done using either the Illumina MiSeqReporter.
Statistical analysis
The association between TP53 mutational status and patients’ demography, such as age, ethnicity, gender, and baseline disease characteristics, such as percentage of bone marrow blasts, white blood cell count, haemoglobin levels and LDH Levels, were analysed using Chi Square and Fisher exact test for categorical variables, and Independent T-test for continuous variables. The clinicopathology of all the TP53-mut cases were described and the correlation of TP53 mutational status with other genetic mutations, such as NPM1, FLT3, ASXL1 and TET2, was also analysed using Fisher exact test.
Association of variables such as age, TP53 mutational status and cytogenetic risk, with treatment response are also analysed using Fisher exact test. Overall survival analysis based on mutational status was done using Kaplan-Meier survival analysis. Univariate and multivariate analysis were used to predict treatment response.
All statistical analysis in this report was performed using IBM® SPSS® Statistics Version 22.
Results
Patient characteristics
The study cohort consisted of a total of 166 AML patients. The median age of the patients was 46.5 years. There are 85 male patients (51.2%) and 81 female patients (48.8%) respectively. TP53 genetic mutations were found in 9 patients (5.4%) with a larger proportion of males (77.8%) than females (22.2%). No TP53 mutations was found in Malay and Indian patients in this study. The various baseline clinical characteristics and molecular abnormalities were compared between the TP53mut and TP53wt groups as summarised in (Table 1). Only cytogenetic risk classification and complex cytogenetics were found to be statistically significant characteristics.
Table 1. Comparison of clinical and disease characteristic according to mutational status.
Baseline characteristic |
Overall
(n=166) |
TP53-mutated
(n=9) |
TP53-wild type
(n=157) |
p Value |
Age (y), mean (range) |
47.36 (17 – 81) |
50.00 (20 – 81) |
47.20 (17 – 80) |
0.608 |
Ethnicity, n (%) |
0.735 |
Chinese |
100 (60.2) |
6 (66.7) |
94 (59.9) |
|
Malay |
18 (10.8) |
0 (0.0) |
18 (11.5) |
|
Indian |
11 (6.6) |
0 (0.0) |
11 (7.0) |
|
Others |
37 (22.3%) |
3 (33.3) |
34 (21.7) |
|
Gender, n (%) |
0.169 |
Male |
85 (51.2) |
7 (77.8) |
78 (49.7) |
|
Female |
81 (48.8) |
2 (22.2) |
79 (50.3) |
|
Bone marrow blasts (%), mean (range)# |
64.66 (1–99) |
76.00 (26–98) |
63.89 (1–99) |
0.202 |
White blood cell count (×109/L)*, mean (range) |
60.58 (0.45–435.00) |
49.76 (0.45–312.70) |
61.16 (0.79–435.00) |
0.714 |
Haemoglobin levels (g/L)**, mean (range) |
8.33 (2.0–14.0) |
8.56 (5.1–11.1) |
8.32 (2.0–14.0) |
0.72 |
Platelet levels (×109/L)***, mean (range) |
65.08 (1–772) |
59.73 (1–178) |
65.37 (5–772) |
0.836 |
LDH levels (U/L), mean (range) **** |
1285.73 (304–5720) |
1601.25 (491–5720) |
1266.90 (304–5033) |
0.36 |
Cytogenetic risk stratification, n (%) ***** |
0.002 |
Favourable |
29 (18.4) |
2 (22.2) |
27 (18.1) |
|
Intermediate |
95 (60.1) |
1 (11.1) |
94 (63.1) |
|
Adverse |
34 (21.5) |
6 (66.7) |
28 (18.8) |
|
Complex Cytogenetics, n (%) |
<0.001 |
Complex |
20 (12.0) |
6 (66.7) |
14 (8.9) |
|
Non-complex |
146 (88.0) |
3 (33.3) |
143 (91.1) |
|
In addition, six out of the nine TP53mut was associated with complex karyotype (p=0.002) and was categorised to have an adverse cytogenetic risk. The frequency of TP53 mutations in cytogenetically complex AML patients was 30% (six out of twenty patients). Of the remaining three TP53mut cases, two had favourable APML (t (15;17)) translocation and one had normal cytogenetics (Table 2). Both APML cases have a white cell count £ 10x109/L and platelet count £ 40x109/L, which are considered to be intermediate risk APML.
Table 2. Summary of the clinicopathology of all the TP53-mut cases.
Case No. |
Age (yr) |
Karyotype |
Other concomitant mutations |
Treatment intent |
Response to induction |
Relapsed/ refractory |
Survival status |
1 |
81 |
Complex |
ASXL1, IDH1 |
Palliative |
NA |
NA |
Dead |
2 |
48 |
t(15;17) |
TET2 |
Curative |
CR |
No |
Alive |
3 |
31 |
Complex |
Nil |
Curative |
No CR (achieved CR after allogeneic SCT) |
Yes |
Dead |
4 |
61 |
Complex |
Nil |
Curative |
No CR |
Yes |
Loss f/u |
5 |
20 |
Complex |
NPM1 |
Curative |
No CR |
Yes |
Dead |
6 |
29 |
t(15;17) |
ASXL1, RUNX1, DNMT3a, TET2 |
Curative |
NA |
NA |
Loss f/u |
7 |
56 |
Complex |
Nil |
Curative |
Induction death |
NA |
Dead |
8 |
54 |
Complex |
Nil |
Curative |
Induction death |
NA |
Dead |
9 |
70 |
Normal |
NPM1, TET2 |
Curative |
Induction death |
NA |
Dead |
Types of TP53 mutations and correlation with other genetic mutations
Of the 9 TP53 mutations, 7 were missense mutations while 2 were nonsense mutations that resulted in premature truncation of the protein (Figure 1).
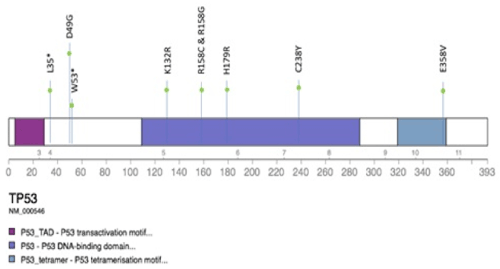
Figure 1. The type and location of TP53 mutations detected
Overall, the most frequent gene mutation among 166 AML patients is NPM1 (22.9%), followed by ASXL1 (20.2%) and RUNX (18.1%) (Table 3). TP53 gene mutation was found in 5.4% of patients. It appears to be mutually exclusive with FLT3 and IDH2 mutation. Two cases with TP53 mutations had concomitant NPM1 mutation, of which one (Case 5) had complex cytogenetics and did not achieve CR, eventually had progressive disease, and the other (Case 9) had normal karyotype but was complicated by induction death. Another two had ASXL1 mutation (Case 1 and 6). TP53 mutation AML appeared to be associated with TET2 mutation (3/9, 33.3%) compared to 10.2% of the TP53wt (p=0.069, Fisher-exact test).
Table 3. Correlation of TP53 mutations with other recurrent genetic mutations.
Gene Mutation |
Overall (n=166) |
Mutated TP53 (n=9) |
Wild-type TP53 (n=157) |
p Value |
NPM1, n (%) |
|
|
|
1 |
Wild type |
128 (77.1) |
7 (77.8) |
121 (77.1) |
|
Mutated |
38 (22.9) |
2 (22.2) |
36 (22.9) |
|
FLT3, n (%) |
|
|
|
0.592 |
Wild type |
133 (80.1) |
9 (100) |
124 (79.0) |
|
Mutated ITD |
25 (15.1) |
0 (0) |
25 (15.9) |
|
TKD |
8 (4.8) |
0 (0) |
8 (5.1) |
|
ASXL1, n (%) |
|
|
|
0.231 |
Wild type |
149 (89.8) |
7 (77.8) |
142 (90.4) |
|
Mutated |
17 (20.2) |
2 (22.2) |
15 (9.6) |
|
TET2, n (%) |
|
|
|
0.069 |
Wild type |
147 (88.6) |
6 (66.7) |
141 (89.8) |
|
Mutated |
19 (11.4) |
3 (33.3) |
16 (10.2) |
|
RUNX, n (%) |
|
|
|
1 |
Wild type |
136 (81.9) |
8 (88.9) |
128 (81.5) |
|
Mutated |
30 (18.1) |
1 (11.1) |
29 (18.5) |
|
IDH1, n (%) |
|
|
|
0.5 |
Wild type |
154 (92.8) |
8 (88.9) |
146 (93.0) |
|
Mutated |
12 (7.2) |
1 (11.1) |
11 (7.0) |
|
IDH2, n(%) |
|
|
|
0.6 |
Wild type |
149 (89.8) |
9 (100.0) |
140 (89.2) |
|
Mutated |
17 (10.2) |
0 (0.0) |
17 (10.8) |
|
Treatment response and overall survival
For the subsequent analysis, 26 patients, who received palliative treatment, and 20 patients who travelled back to their home country for treatment and are therefore lost to follow-up are excluded from analysis. 11 APML (M3) patients are also excluded from analysis as APML is a subtype of AML with exceptionally good outcomes due to its responsiveness to all-trans retinoic acid (ATRA) therapy that is not representative of other AML subtypes. A total of 109 non-APML patients received curative treatment. The median follow-up time is 52.4 months (95% CI: 35.6 – 69.3).
Six out of seven non-APML TP53-mut patients underwent curative treatment. Three (Cases 7, 8 and 9) were complicated by induction deaths, two (Cases 4 and 5) did not achieve complete remission and had primary refractory disease and the remaining one (Case 3) achieved CR only after allogeneic stem cell transplantation. The patient then relapsed after a short CR of 6.8 months. Compared to wt-TP53 patients, mut-TP53 patients have a much lower CR rate (16.7% vs 82.7%, p<0.005, Fisher exact test) (Table 4). In addition, older age as well as adverse cytogenetics were also associated with lower CR rate.
Table 4. Association between individual risk factors and CR.
Variables |
Achieved CR |
Did not achieve CR |
p Value |
Age, n (%) |
|
|
0.001 |
|
79 (84.0) |
15 (16.0) |
|
≤60 years |
|
|
|
|
4 (36.4) |
7 (63.3) |
|
>60 years |
|
|
|
TP53, n (%) |
|
|
0.001 |
Mutated |
1 (16.7) |
5 (83.3) |
|
Wildtype |
82 (82.8) |
17 (17.2) |
|
Cytogenetic Risk, n (%) |
|
|
0.041 |
Favourable |
11 (84.6) |
2 (15.4) |
|
Intermediate |
53 (84.1) |
10 (15.9) |
|
Adverse |
13 (59.1) |
9 (40.9) |
|
Table 5. Multivariate Cox Proportional Hazards Regression analysis for overall survival.
Risk Factors |
Adjusted HR |
95% CI |
p value |
Mutational Status:
Mut-TP53 vs wt-TP53 |
43.07 |
6.81 – 272.41 |
<0.001 |
Cytogenetic Risk |
|
|
|
Adverse vs Favourable |
1.75 |
0.43 – 7.19 |
0.437 |
Intermediate vs Favourable |
1.35 |
0.37 – 4.96 |
0.648 |
White Blood Count |
1 |
1.00 – 1.01 |
0.175 |
LDH Levels |
1 |
1.00 – 1.00 |
0.364 |
Bone Marrow Blasts Percentage |
1.02 |
1.00 – 1.04 |
0.095 |
TP53 mutation AML was significantly associated with inferior OS compared to TP53wt (p<0.001). Median overall survival (OS) was 0.8 months (95% CI: 0.159-1.484) and 36.8 months (95% CI: 0.000-92.676) respectively (Figure 2). TP53 mutational status remains an independent predictor of OS in multivariate analysis that include cytogenetic risk, WBC, LDH and BM blasts (HR 43.1, 95%CI: 6.8 – 272.4, p<0.001) (Table 4).
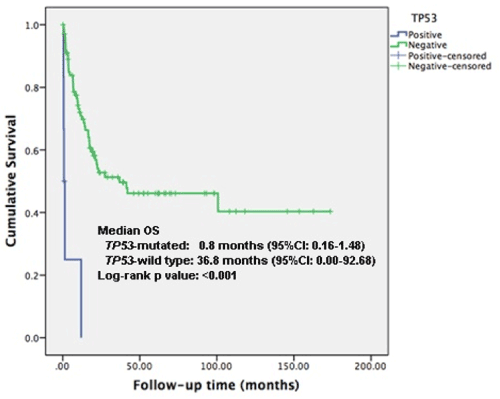
Figure 2. Overall survival according to mutational status
Discussion
2021 Copyright OAT. All rights reserv
Patient characteristics
In our study, we found that the incidence of TP53 genetic mutations in a South East Asian cohort of patients to be 5.4%, which is consistent with previously reported 5 to 10% incidence in various studies in other populations [10]. There appears to be a higher incidence of TP53 mutations in male patients.
Compared with wt-TP53 cohort, mut-TP53 patients are significantly more likely to have a complex karyotype, 66.7% as compared to 8.9% for wt-TP53 patients. Many studies have reported this association, but it remains unclear whether the TP53 genetic mutation and the resultant loss of its tumour suppressor function led to the chromosomal instability or if the TP53 genetic mutation arose secondary to the chromosomal instability. Interestingly, in our cohort of 9 patients, we found 6 cases of mut-TP53 patients with complex cytogenetics, 2 cases of APML mut-TP53 patients as well as a case with normal cytogenetics (Table 2). Further studies are required to determine the causal relationship and elucidate the underlying pathological mechanisms.
While TP53 mutations is associated with an adverse prognosis, one interesting observation in this study is that the mutational status of TP53 did not appear to influence the outcome for APML patients, a subset of AML with exceptionally favourable prognosis due to its sensitivity to all-trans retinoic acid (ATRA) treatment. The two TP53-mut APML patients are classified into the intermediate risk category with a white blood cell count < 10,000/μL and a platelet count < 40,000/μL. One of the APML patient (Case 2) underwent treatment achieved and remained in complete remission after 4 years. The other APML patient (Case 6) returned to his home country for treatment and was lost to follow up.
Types of TP53 mutations and correlation with other genetic mutations
TP53 (tumour protein 53) is a transcription factor composed of a transcription activation domain, a DNA-binding domain, a proline-rich domain, and a tetramerization domain [15]. Most studies have focused on mutations in the DNA-binding domain of TP53 gene. However, recently, Terada et al. reported that TP53 mutations outside the DNA binding domain contributes to poorer outcomes compared to TP53 mutation in the DNA binding domain [16]. In addition, they found that all cases with gene deletion involved a monoallelic deletion with gene mutation on opposite side. The impact of various TP53 mutations and deletion on clinical outcomes is an area that can be further expounded on future studies.
Our study also found a mutual exclusivity of FLT3 and TP53 mutations. Although it did not reach statistical significance, this finding has been corroborated by many studies. FLT3 is a commonly mutated gene in AML patients (21.0% in this study) and FLT3-ITD mutation has been shown to be associated with poor prognosis in patients with normal karyotype and is believed to be a driver mutation in leukemogenesis [17].
In addition, there appears to be a correlation between TET2 and TP53 mutations, which did not reach statistical significance, likely attributable to the small sample size of the study. This is probably the first time that this association has been suggested. Previously other studies have suggested that DNA-methylation regulatory gene mutations, such as DNMT3A and TET2, appear to be founder gene mutations [18,19] and are frequently co-present with high-frequency mutations such as FLT3-ITD and NPM1. Since TP53 mutations tend to be mutually exclusive with FLT3-ITD and NPM1 [20], they are less likely to be associated with DNMT3A and TET2 too.
Treatment response and overall survival
In a multivariate analysis that includes age, cytogenetic risk stratification, WBC, LDH and BM blasts, TP53 mutational status remains an independent predictor of CR and OS. TP53-mut patients are more likely to be resistant to the standard induction chemotherapy and present with primary refractory disease. Hence, they may benefit from allogeneic stem cell transplantation (SCT) or earlier enrolment into clinical trials. Allogeneic SCT could improve the leukemia free survival to around 25% in patients with TP53 who were transplanted in CR1 [21]. Ohgami also reported that cases that received hematopoietic stem cell transplant in the first remission period tended to have a higher rate of OS (p=0.0606) while there was no significant positive outcome for patients that were transplanted not in the first remission [22]. For those who are unfit for allogeneic SCT, treatment with hypomethylating agent like decitabine might produce a better response compared to conventional chemotherapy [23].
There has also been an increasing interest in small molecule target drugs. Selinexor, a protein pump inhibitor, is one such candidate. It works by binding with and inhibiting the nuclear export protein XPO1 (also called CRM1), leading to the accumulation of tumor suppressor proteins such as p53 in the cell nucleus. This reactivates and enhances tumour suppressor function. Preclinical studies have suggested that XPO1 inhibitors lead to the targeted apoptosis of cancer cells, while sparing normal cells [24]. This may be a promising candidate drug for patients with TP53 mutations and relapsed AML patients, whereby the incidence of TP53 mutations may be high.
There has been increasing interest in targeted therapies based on genetic mutations. As missense TP53 mutations are most common and this often results in the loss of DNA binding ability and accumulation of mutated p53 protein in the tumour cells at high concentration, the reactivation of these mutated p53 protein hence holds promising therapeutic potential [25] Various drugs/ compounds have been studied. Of note, PRIMA-1 and PRIMA-1MET/APR-246 have been validated in animal models and is currently in clinical trials [26]. Successful development of drugs targeting TP53 mutations will help broaden the range of therapeutic options available for patients as well as potentially reverse the poor prognosis associated with TP53 mutations.
Conclusion
Our results in the South East Asian study cohort affirmed the extremely dismal prognosis of TP53 mutation AML, in line with the European Leukemia Net guidelines. TP53-mut patients are significantly more likely to be associated with complex karyotype, lower CR rates and shorter OS. TP53 mutation testing should be included as part of the pre-chemotherapy workout to improve risk stratification of AML. However, the role of TP53 prognostication in APML remains to be explored as it does not appear to be affecting the prognosis. The ability to draw correlations between TP53 and other recurrent genetic mutations is limited by the small patient cohort in this study. The significance of TP53 mutation association with TET2 mutation may be worth further exploration in a larger study cohort. In addition, genetic testing for TP53 should be done at various time points in treatment to elucidate the contribution of TP53 in the pathogenesis, clonal evolution and development of treatment resistance.
References
- Ilyas AM, Ahmad S, Faheem M, Naseer MI, Kumosani TA, et al. (2015) Next generation sequencing of acute myeloid leukemia: influencing prognosis. BMC Genomics 16: S5. [Crossref]
- De Kouchkovsky I, Abdul-Hay M (2016) 'Acute myeloid leukemia: a comprehensive review and 2016 update'. Blood Cancer J 6: e441. [Crossref]
- Prokocimer M, Molchadsky A, Rotter V (2017) Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: projections on diagnostic workup and therapy. Blood 130: 699-712.
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, et al. (2009) The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 114: 937-951. [Crossref]
- Zaidi SZ, Owaidah T, Al Sharif F, Y. Ahmed S, Chaudhri N, et al. (2008) The challenge of risk stratification in acute myeloid leukemia with normal karyotype. Hematol Oncol Stem Cell Ther 1: 141-158.
- Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis E (2013) The next-generation sequencing revolution and its impact on genomics. Cell 155: 27-38. [Crossref]
- Dohner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, et al. (2010) Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 115: 453-474. [Crossref]
- Levine AJ, Oren M (2009) The first 30 years of p53: growing ever more complex. Nat Rev Cancer 9: 749-758. [Crossref]
- Yan W, Zhang Y, Zhang J, Liu S, Cho SJ, et al. (2011) Mutant p53 protein is targeted by arsenic for degradation and plays a role in arsenic-mediated growth suppression. J Biol Chem 286: 17478-1786.
- Kadia TM, Jain P, Ravandi F, Garcia-Manero G, Andreef M, et al. (2016) TP53 mutations in newly diagnosed acute myeloid leukemia: Clinicomolecular characteristics, response to therapy, and outcomes. Cancer. [Crossref]
- Bowen D, Groves MJ, Burnett AK, Patel Y, Allen C, et al. (2009) TP53 gene mutation is frequent in patients with acute myeloid leukemia and complex karyotype, and is associated with very poor prognosis. Leukemia 23: 203-206. [Crossref]
- Seifert H, Thiede C (2009) The prognostic impact of 17p (p53) deletion in 2272 adults with acute myeloid leukemia. Leukemia 23: 656-663.
- Kihara R, Nagata Y, Kiyoi H, Kato T, Yamamoto E, et al. (2014) Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia. 28: 1586-1595. [Crossref]
- Tan M, K S Ng I, Chen Z, Ban K, Chiu L, et al. (2017) Clinical implications of DNMT3A mutations in a Southeast Asian cohort of acute myeloid leukaemia patients. J Clin Pathol 70: 669-676.
- Joerger AC, Fersht AR (2010) The tumor suppressor p53: from structures to drug discovery. Cold Spring Harb Perspect Biol 2: a000919.
- Terada K, Yamaguchi H, Ueki T, Usuki K, Kobayashi Y, et al. (2018) Full-length mutation search of the TP53 gene in acute myeloid leukemia has increased significance as a prognostic factor. Ann Hematol 97: 51-61.
- Testa U, Pelosi E (2013) The Impact of FLT3 Mutations on the Development of Acute Myeloid Leukemias. Leuk Res Treatment 2013: 275760. [Crossref]
- O'Brien EC, Brewin J, Chevassut T (2014) DNMT3A: the DioNysian MonsTer of acute myeloid leukaemia. Ther Adv Hematol 5: 187-96.
- Ganguly BB, Kadam NN (2016) Mutations of myelodysplastic syndromes (MDS): An update. Mutat Res Rev Mutat Res 769: 47-62.
- Hou HA, Chou WC, Kuo YY, Liu CY, Lin LI, et al. (2015) TP53 mutations in de novo acute myeloid leukemia patients: longitudinal follow-ups show the mutation is stable during disease evolution. Blood Cancer J 5: e331. [Crossref]
- Poire X, Labopin M, Maertens J, Yakoub-Agha I, Blaise D, et al. (2017) Allogeneic stem cell transplantation in adult patients with acute myeloid leukaemia and 17p abnormalities in first complete remission: a study from the Acute Leukemia Working Party (ALWP) of the European Society for Blood and Marrow Transplantation (EBMT). J Hematol Oncol 10: 20.
- Ohgami RS, Ma L, Merker JD, Arber D (2015) Next-generation sequencing of acute myeloid leukemia identifies the significance of TP53, U2AF1, ASXL1, and TET2 mutations. Mod Pathol 28: 706-714.
- Welch JS, Petti AA, Miller CA, Fronick CC, O’Laughlin M, et al. (2016) TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N Engl J Med 375: 2023-2036.
- Etchin J (2013) KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. Br J Haematol 161: 117-127.
- Oren M, Tal P, Rotter V (2016) Targeting mutant p53 for cancer therapy. Aging (Albany NY) 8: 1159-1160.
- Parrales A, Iwakuma T (2015) Targeting Oncogenic Mutant p53 for Cancer Therapy. Front Oncol 5: 288. [Crossref]


