Abstract
The present study conducted proteomic analysis to identify ovarian proteins induced by GnRH/GnIH in comparison with untreated control mice. We used methods of 2-dimentional gel electrophoresis and LC-MS/MS for protein identification. Out of the 25 differentially expressed spots from each of the GnRH/GnIH treated ovaries we were randomly selected 18 protein spots identified by LC-MS/MS. The proteins 14-3-3, prohibitin and superoxide dismutase (SOD1) identified by LC-MS/MS were further verified by Western blotting, which confirmed that GnRH/GnIH-induced changes in expression of 14-3-3, prohibitin and SOD1 were similar to the observations of proteomics. These selected proteins, 14-3-3, prohibitin and SOD1, were further have been the correlation with various ovarian activities, such as cell proliferation, survival or markers of apoptotic. Out of 18 identified GnRH-induced proteins, 9 showed up-regulations whereas other 9 showed down-regulations. This finding suggests that GnRH acts both by stimulating and inhibiting expression of protein. Interestingly, in the GnIH-induced ovary, 15 out of 18 proteins were significantly down-regulated suggesting that, GnIH acts mainly by inhibiting the expression of proteins involved in ongoing ovarian activities. Thus, the present study indicates many of the identified GnRH/GnIH-induced proteins are potentially linked to ovarian follicular development, atresia, steroidogenesis and cancer.
Key words
gonadotropin releasing hormone (gnrh); gonadotropin inhibitory hormone (gnih); rf-amide related peptide-3 (rfrp-3); two-dimensional gel electrophoresis; apoptosis; ovarian cancer.
Introduction
The decapeptide gonadotrophin releasing hormone (GnRH) or its agonist (GnRH-Ag), a major regulatory neuropeptide first isolated from the mammalian hypothalamus, is involved in primary control of reproductive processes across vertebrates [1-3]. The presence of GnRH and GnRH-receptor in the ovary of several mammalian species, including human, strongly suggests the local synthesis and biological actions of GnRH in the ovary as well as hypophysiotrophic actions [4-10].
GnRH has both stimulatory and inhibitory actions on various ovarian activities. In the ovaries of rats and mice, several studies have reported that significant variations of the expressions of GnRH and GnRH receptor during the estrous cycle [9,11]. GnRH exerts a stimulatory action on preovulatory follicles by inducing oocyte maturation [12] and follicle rupture [13]. In smaller follicles, GnRH exerts an inhibitory action on the induction of gonadotropin-receptor [14]. The increased concentration of GnRH during proestrus and estrus phases suggests possible involvement of this neuropeptide for the selection of follicles for growth, differentiation and atresia in the ovary [9,11]. The presence of GnRH and its receptor in thecal cells of antral follicles suggests possible involvement of this neuropeptide in regulation of gonadal steroidogenesis [8,15]. The presence of GnRH and its receptor in the granulosa cells of atretic follicles suggests a role of GnRH in the induction of follicular atretia in the ovary of mice [9]. The treatment of GnRH agonist in vivo has been shown to reduce proliferative activity and induce apoptosis in granulosa cells [16]. The anti-gonadotropic effect of GnRH was found to be mediated by down regulation of the expression of LH- and FSH-receptors and suppression of the expression of steroidogenic enzymes in the rat ovary [9,17,18]. Furthermore, it has been suggested that GnRH may have permissive actions on the process of luteal regression [19]. The mRNA expression of GnRH receptor was also found in luteinized granulosa cells in human [8]. However, not much is known about the mode of actions of GnRH to regulate various physiological actions in the mammalian ovary.
Another neuropeptide, the mammalian GnIH peptide, RF-amide related peptide-3 (RFRP-3) and its receptor have been demonstrated in the ovary of rhesus monkey and mice [20,21]. Extensive studies have described that GnIH is also an important intra-ovarian factor inhibiting ovarian follicular development and steroidogenesis [22,23]. Previous studies identified two G-protein couple receptors, GPR 147 and GPR 74, as the GnIH-receptor [24,25]. The GnIH-receptor is localized in the theca and granulosa cells, with significantly lower level in preovulatory follicle as compared to the smaller growing follicles [9]. The relative abundance of both GnRH and GnIH mainly in the antral follicles of mice during proestrus and estrus phases suggests the possible involvement of these neuropeptides in the selection of follicles for growth and atresia. The expressions of both GnRH and GnIH in the ovary of mice suggest the possibility for the interaction between these peptides [9]. Our earlier study has demonstrated a positive correlation between the levels of GnRH and GnIH in the gonad from birth to senescence [26]. It has further been found that GnIH usually inhibits the secretion and action of GnRH [21,22].
The studies described so far suggest that both GnRH and GnIH act as important autocrine/paracrine factors that modulate follicle development, apoptosis, luteinization and steroidogenesis in the ovaries of mammals. GnRH and GnIH have also been proposed as a possible treatment for variety of reproductive dysfunctions [10,27]. Our recent study suggests that GnRH may restore ovulation in polycystic ovarian (PCO-mice) and thus may be utilized as a therapeutic agent to treat polycystic ovarian syndrome [10]. GnRH may also be useful in the treatment of hypogonadotropic hypogonadism and suppression of fertility. Because GnIH is the first identified neuropeptide that has a direct inhibitory action on gonadal functions [20,21], GnIH may develop as an anti-fertility agent [10]. However, many of the functions of GnRH and GnIH in the ovary are still unclear. Specific molecular changes occurred in the ovary in response to the treatment with either GnRH/GnIH and may provide knowledge about unknown processes of the modulation of various ovarian activities by these neuropeptides. To identify ovarian proteins whose expressions, change (induced or repressed) in response to the treatment with either GnRH or GnIH, the present study employed a proteomic approach comprised of two-dimension (2D) gel electrophoresis and LC-MS/MS. To verify the proteomic data, the expression levels of selected proteins were evaluated by Western blot analysis in the mice ovary.
Material and methods
Antibodies and reagents
Chemicals and antibodies: Mouse RFRP-3 long form (VNMEAGTRSHFPSLPQRF-NH2), a mouse GnIH ortholog, was kindly provided by Prof. Tsutsui, Department of Biology, Waseda University, Tokyo, Japan [28]. GnRH-Ag used in the study was ([DTrp6, Pro9-NEt] GnRH). It was kindly provided by Dr. Marvin Karten, Contraceptive branch, NIH, USA. All the general chemicals used in this study were purchased from Merck, New Delhi, India. Detail of antibody used was given in Table 1.
Table 1. Details of antibodies used for western blot
Antibody |
Target species |
Species raised in Monoclonal/ Polyclonal |
Source |
Concentration (used for Western blotting |
Prohibitin |
Human |
Rabbit; Polyclonal |
GenScript (A0070640) |
1:1500 |
SOD1
|
Human |
Rabbit; Polyclonal |
GenScript (A0100540) |
1:3000 |
PARP-1 |
Human |
Rabbit; Polyclonal |
Santa cruz Biotechnology Inc (H-250,SC 7150) |
1:1000 |
Caspase-3 |
Human |
Rabbit; Polyclonal |
Santa cruz Biotechnology
Inc. (H-277, SC 7148) |
1:1000 |
PCNA
|
Human |
Rabbit; Polyclonal |
Thermo Fisher Scientific,
Rockford, USA, PA1-38424 |
1:1500 |
BCl2 |
Human |
Rabbit; Polyclonal |
Santa cruz Biotechnology Inc. (H-61, SC 7938) |
1:1500 |
Actin |
β-Actin |
Mouse; Monoclonal |
Sigma A2228, 128K4813 |
1:20,000 |
Secondary |
Rabbit |
Goat; Polyclonal |
Bio Geni |
1:4000 |
Animals
All the experiments were conducted in accordance with principles and procedures approved by the Departmental Research Committee of Banaras Hindu University, India. Mice (Mus musculus) of Parkes Strain were housed under constant condition of temperature (24±2 ̊C) and humidity in a photoperiodically controlled room (12-h light, 12-h dark) of our animal house and were provided with commercial food (Pashu Aahar Kendra, Varanasi, India) and tap water ad libitum. Adult (12 weeks old) female mice of about similar body weight and exhibiting at least two consecutives 4 to 5 days cycles were used in this experiment. Vaginal cytology was observed early in the morning to check the proestrus phase of cycling mice. Vaginal cytology showing presence of nucleated epithelial cells confirms the proestrus phase of cycling mice [29]. Mice with regular estrus cycles were randomly allocated into three groups.
Experimental design
In order to execute the experiments mice ovaries (n=30) were immediately dissected out at proestrus phase of estrus cycle and subjected to in vitro culture for 24 h at 37 ̊C in humified atmosphere 5% CO2. Ovaries were divided into three groups: GnIH (100 ng/ml), GnRH-Ag (100 ng/ml) and control group (only media). Ovaries were pooled out and were washed with PBS thrice and stored at −80 °C until used for 2-DE and immunoblotting where as media was used for hormone assay. The experiments were repeated thrice to get same results. After the two-dimensional gel electrophoresis spots from 2-DE gel of two groups compared with control were analyzed with PDQuest image analysis software. Spots exhibiting greater than two-fold change in expression as determined by Student t-test (p<0.05) were selected for protein identification by tandem mass spectrometry. Out of 25 differentially expressed spots 18 spots were selected using non-targeted approach from each group for LC-MS/MS analysis.
Validation of proteomic study
For the validation of the proteomics study, one of the protein spots identified by LC–MS/MS analysis was confirmed by Western blot analysis. The protein 14-3-3 zeta/delta was selected for further study because it is involved in regulation of diverse cellular processes such as oxidative stress, inflammation, apoptosis, cell proliferation and survival [30]. 14-3-3 might be involved in various physiological changes occurring in the mice ovary. Other than 14-3-3 zeta/delta we have also selected prohibitin and SOD1 to see the changes occurring in the ovary in order to further validation of our results. Beside this, there were presence of apoptotic markers and cell proliferation marker in the ovaries because ovarian granulosa cells showed atretia. Therefore, in order to examine changes occurring in ovarian cells this study was undertaken to verify whether GnRH and GnIH were regulating the expression of apoptotic markers (Caspase-3 and PARP-1) and survival factors (BCl2 and PCNA). To achieve this immunoblotting of apoptotic markers and cell proliferation marker was conducted in the mice ovaries collected during the proestrus phase of the estrus cycle. Following this, correlation study was undertaken to find out how 14-3-3, prohibitin and sod1 with apoptotic and cell survival marker proteins regulate the ovarian physiology. To achieve this, the mice ovaries collected after in vitro culture were processed for immunoblot analysis of 14-3-3 zeta/delta, Prohibitin, sod1, caspase-3, PARP-1, PCNA and BCl2 proteins. The relative concentration of the proteins in ovaries was obtained by densitometric analysis of western blots. The correlative changes in the concentrations of 14-3-3 zeta/delta protein, prohibitin and SOD1, together with of apoptotic marker (caspase-3, PARP-1), the changes in the concentrations cell proliferative (PCNA) marker and cell survival marker (BCl2) were analyzed.
In vitro study
Proestrus ovaries were used for in vitro culture because they contain many antral follicles and to maintain uniformity in the stage of ovaries used in all the groups. Adult female mice (n=30) were killed by decapitation under mild dose of anesthetic ether. These ovaries were quickly dissected out and cleaned from any adhered fat tissue and oviduct in medium Dulbecco Modified Eagle’s Medium (DMEM; Himedia, Mumbai, India) containing penicillin (250 U/ml) and streptomycin sulfate (250 μg/ml). Ovaries were cultured by the method as described previously [22,31]. Culture media was a mixture of DMEM (with sodium pyruvate and L-glutamine) and Ham’s F-12 (1:1; v: v) (Himedia, Mumbai, India) containing penicillin (100 U/ ml), streptomycin sulfate (100 μg/ml) and BSA (0.1% w/v, Sigma Chemicals Co., St Louis, USA). Intact ovaries (one per well in culture plate) were cultured in 1 ml medium in a humidified atmosphere with 95% air and 5% CO2 to maintain pH 7.4 for 24 h at 37°C. The doses of neuropeptides were selected on the basis of studies conducted previously in our laboratory [22,31]. The treatment of GnIH (RFRP-3 LF) 100 ng/ml, GnRH 100 ng/ml and control (without any neuropeptide) was given in a total volume of 10 μl/ml medium per well of culture plate. Ovaries pooled out after culture, washed several times with PBS and kept frozen -80°C for 2-Dimensional gel electrophoresis and Western blot analysis. The culture media was kept for hormone assay. Each treatment group was run in triplicate with the same results.
Two-dimensional gel electrophoresis
Ovaries were collected after in vitro culture, homogenized by sonication in ice cold 1 M Tris (pH 7.6) having 1µg/ml aprotinin, 100 µg/ml PMSF. Homogenized protein was precipitated in ice cold acetone (sample: acetone; 1:10) for overnight at -20 ºC and then centrifuged at 10,000 rpm for 5 minutes at 4°C. Acetone was discarded after centrifugation and pellet was allowed to dry until smell of acetone goes off. The pellet was then resuspended in rehydration buffer (7M urea, 2M thiourea, 4% CHAPS, 50 mM DTT and 0.2% ampholites). Protein estimation was performed by Bradford Assay. Strips (pH 4-7, 11cm) were rehydrated passively for 16 hours and then IEF was performed using a PROTEAN IEF cell (Bio-Rad) for 40,000 V-h. After completion of IEF, the strips were equilibrated in two base buffers (50 mM Tris/Cl, 2% (v/v) SDS, 6 M urea, and 20% glycerol) containing first 1.4 M diothiothreitol (DTT) and then 1.3 M iodoacetamide. The second dimension was run onto 12 % SDS-PAGE. The gel was washed thrice for 15 min prior to staining with CBBG-250. For quantification purposes, the stained gels were scanned (GS- 800 Densitometer, Bio-Rad) and analyzed with PDQuest image analysis software (Bio-Rad) followed by LC-MS/MS. Each treatment group was run in triplicate with the same results.
LC-MS/MS from 2D gel spots
Out of 25 spots, 18 Spots from the 2 D gel were randomly selected for the identification by LC-MS/MS using a Thermo-Finning LTQ mass spectrometer coupled to a Multimate 3000 nanoflow HPLC. Spots from 2D gels were typically stored in clean (rinsed with mass spec grade Acetonitrile) Eppendorf tubes for transportation or storage. Spots (gel plugs) were washed three times in 100% acetonitrile for at least 10 min per wash to remove any stain. After the last wash the spots were placed in a Speed-Vac and dried for 20 min. The dried spots were rehydrated in trypsin digest buffer (50 mM ammonium bicarbonate; Am Bic). Spots were first reduced in diothiothreitol (DTT) 10 mM at 56 ̊C for at least 30 min and alkylated with 15 mM Iodoacetic Acid for 30 min at room temperature in the dark. Samples were then digested with mass spec grade trypsin 20 ng/µl for 4 hrs at 37 ̊C. Just before analysis, the sample was acidified by the addition of Formic acid to 0.1% [32].
Spots from 2D gels were analyzed using a 60 min LC/MS run on a Multimate 3000 nanoflow HPLC. The sample was loaded onto a trap (C18 PepMap 100, 300µm X 5 mm, Thermo) at 5 µl/min for 5 min. The multiport valve was then switched and the trap was eluted onto a reverse phase column using a gradient of acetonitrile in water 5%-30% over ten min at a flow rate of 200 nl/min. Eluate from the column was directly sprayed into an LTQ ion trap mass spectrometer using nanospray (Michrom). Spectra are collected using X calibur 2.2 software (Thermo) using a threshold setting of 200. Spectra were searched against a mouse Swiss-Prot database using Proteome Discoverer 3.1 software (Thermo). The percolation selection algorithm was used, set to a 5% false discovery rate to select the best database-hits (Tables 2 and 3).
Table 2. Details of spot analysis: GnIH treated groups
|
|
Regulation of protein |
Accession No. |
|
|
|
Proteins |
Unique Proteins |
Pep-tides |
PSMs |
AAs |
Molecular wt. (KDa) |
Cal. pI |
-
|
|
-
-
B1AQX9 |
Protein N4bp2(fragment)OS=Mus musculus GN=N4bp2PE=4SV=2-[ E9Q9B2_MOUSE]
MCG124046 OS= Mus musculus GN=Prss 1PE=2 SV=1-[ Q9Z1R9_MOUSE]
SRC kinase-signalling inhibitor 1 OS=Mus musculus GN=Srcin 1 PE=1SV=1-[ B1AQX9_MOUSE] |
-
-
-
|
-
-
-
|
2
1
5 |
1
1
2 |
1
1
2 |
13
3
15 |
1498
246
1217 |
-
-
-
|
-
-
-
|
-
|
|
-
B1AQX9
E9Q9B2 |
Isoform 2 of Tropomyosin beta chain OS= Mus musculus
SRC kinase-signalling inhibitor 1 OS=Mus musculus GN=Srcin 1 PE=1SV=1-[ B1AQX9_MOUSE]
Protein N4bp2(Fragment)OS
|
-
-
-
|
-
-
-
|
16
6
2 |
6
3
2 |
6
3
2 |
7
6
6 |
284
1174
1498 |
-
-
-
|
-
-
-
|
-
|
|
-
-
-
|
Tropomyosin alpha-1 chain OS=Mus musculus GN=Tpm 1PE=3SV=1-[E9Q450_MOUSE]
-
Transmembrane protein serine 11F OS=Mus musculus GN=Tmprss 11fPE=SV=1-[TM11F_MOUSE]
|
-
-
-
|
-
-
-
|
28
1
1 |
9
1
1 |
16
1
2 |
29
4
3 |
284
246
439 |
-
-
-
|
-
-
-
|
-
|
|
-
-
-
-
-
-
-
-
|
-
Isoform 2 of Telomere repeats binding bouquet formation protein 1 OS= Mus musculus GN=Ccdc79-
-
14-3-3 PROTEIN EPSILON
Interleukin-1 receptor- associated kinase 4OS= Mus musculus GN=Irak4 PE=2SV=1-[ Q3USX5_MOUSE]
Protein N4bp2(Fragment)OS
Isoform 2 of Ras-responsive element-binding protein 1OS= Mus musculus GN=Rreb1-[RREB1_MOUSE]
E3 SUMO-protein ligase RanBP2 OS= Mus musculus GN=Ranbp 2PE=1 SV=2-[RBP2_MOUSE]
Protein Armcx4 OS= Mus musculus GN= Armcx4PE=4 SV=1-[ E9PWM3_ MOUSE] |
-
-
-
-
-
-
-
-
|
-
-
-
-
-
-
-
-
|
1
4
2
2
2
5
1
1 |
1
1
1
3
3
4
5
4 |
1
2
1
3
4
4
6
6 |
3
4
3
5
12
8
9
8 |
246
705
130
453
1498
1618
3053
2356 |
-
-
-
-
-
-
-
-
|
-
-
-
-
-
-
-
-
|
-
|
|
-
-
-
-
-
-
-
|
14-3-3 protein zeta/delta (Fragments)OS=GN=Ywhaz PE=4 SV=1-[ D3YXN6_ MOUSE]
-
14-3-3 protein sigma OS
Isoform2 of 14-3-3 protein theta OS
Cadherin-7 OS=Mus musculus GN=Cdh7PE
Isoform2 of ProteinRUFY3 OS
Troponin I, slow skeletal muscle (Fragment)OS |
-
-
-
-
-
-
-
|
-
-
-
-
-
-
-
|
4
1
1
4
1
4
3 |
5
1
1
3
2
4
1 |
5
1
3
3
2
4
1
|
15
3
7
4
4
5
2 |
88
246
248
243
785
665
60 |
-
-
-
-
-
-
-
|
-
-
-
-
-
-
-
|
-
|
|
-
|
Adrenodoxin, mitochondrial
|
-
|
-
|
1 |
1 |
1 |
2 |
188 |
-
|
-
|
-
|
|
-
|
Tropomyosin alpha-1 chain |
-
|
-
|
28 |
2 |
2 |
2 |
245 |
-
|
-
|
-
|
|
-
-
|
-
Annexin A5 |
-
-
|
-
-
|
1
1 |
1
2 |
1
2 |
2
3
|
246
319 |
-
-
|
-
-
|
-
|
|
-
-
|
-
Phosphatidylethanolamin-binding protein 1OS |
-
-
|
-
-
|
1
2 |
1
5 |
1
5 |
6
8 |
246
187 |
-
-
|
-
-
|
-
|
|
-
-
|
Cytochrome oxidase subunit 5A, mitochondrial OS
|
-
-
|
-
-
|
1
1 |
5
2
|
5
2 |
9
4 |
135
146 |
-
-
|
-
-
|
-
|
|
-
-
-
-
-
-
|
-
Isoform 3 of Dynamin-1 OS
Isoform 3 of Telomere repeats-binding bouquet formation protein 1 OS
Isoform 4 of Telomere repeats-binding bouquet formation protein 1 OS
Telomere repeats binding bouquet formation protein 1 OS
Isoform 2 of Telomere repeats-binding bouquet formation protein 1 OS
|
-
-
-
-
-
-
|
-
-
-
-
-
-
|
1
1
1
1
1
1 |
1
0
0
0
0
0 |
1
1
1
1
1
1 |
2
2
1
1
1
1 |
246
851
250
249
768
705 |
-
-
-
-
-
-
|
-
-
-
-
-
-
|
-
|
Downregulation (absent in treatment group) |
-
-
-
-
-
|
-
Isoform 3 of Telomere repeats-binding bouquet formation protein 1 OS
Isoform 4 of Telomere repeats-binding bouquet formation protein 1 OS
Telomere repeats binding bouquet formation protein 1 OS
Isoform 2 of Telomere repeats-binding bouquet formation protein 1 OS |
-
-
-
-
-
|
-
-
-
-
-
|
1
1
1
1
1 |
1
0
0
0
0 |
1
1
1
1
1 |
2
2
2
2
2 |
246
250
249
768
705 |
-
-
-
-
-
|
-
-
-
-
-
|
-
|
|
-
-
-
-
-
|
-
Isoform 3 of Telomere repeats-binding bouquet formation protein 1 OS
Isoform 4 of Telomere repeats-binding bouquet formation protein 1 OS
Telomere repeats binding bouquet formation protein 1 OS
Isoform 2 of Telomere repeats-binding bouquet formation protein 1 OS |
-
-
-
-
-
|
-
-
-
-
-
|
1
1
1
1
1 |
0
0
0
0
0 |
1
1
1
1
1 |
2
3
2
2
2 |
246
250
249
768
705 |
-
-
-
-
-
|
-
-
-
-
-
|
-
|
|
-
-
-
|
-
Isoform 2 of AT-rich interactive domain-containing protein
AT-rich interactive domain-containing protein 4B OS |
-
-
-
|
-
-
-
|
1
1
1 |
0
0
0 |
1
1
1 |
2
2
2 |
246
1227
1314 |
-
-
-
|
-
-
-
|
-
|
|
-
-
-
|
-
Homeobox protein engrailed-2 OS
Chromosome-associated kinesin KIF4 OS |
-
-
-
|
-
-
-
|
1
1
1
|
0
0
0 |
1
1
1 |
2
1
1 |
246
324
1231 |
-
-
-
|
-
-
-
|
-
|
|
-
-
-
-
-
-
|
Isoform 3 of Telomere repeats-binding bouquet protein1
Isoform 4 of Telomere repeats-binding bouquet protein1
Telomere repeats-binding bouquet protein1
Isoform 2 of Telomere repeats-binding bouquet protein1
-
Chromosome-associated kinesin KIF4 OS |
-
-
-
-
-
-
|
-
-
-
-
-
-
|
1
1
1
1
1
1 |
0
0
0
0
0
0 |
1
1
1
1
1
1 |
1
1
1
1
1
1 |
250
249
768
705
246
1231 |
-
-
-
-
-
-
|
-
-
-
-
-
-
|
-
|
|
-
-
-
-
-
-
|
-
MCG15083 OS
Protein Armcx4 OS
Nesprin-2 OS
Neurogenic locus notch homologue protein 3OS
Protein Vmn1r82 OS |
-
-
-
-
-
-
|
-
-
-
-
-
-
|
1
2
1
6
3
1 |
2
1
5
6
3
1 |
2
2
7
6
5
1 |
9
6
12
8
7
2 |
246
246
2356
6870
1964
304 |
-
-
-
-
-
-
|
-
-
-
-
-
-
|
-
|
|
-
-
-
-
-
-
-
|
-
-
Telomere repeats-binding bouquet formation protein1
Isoform 3 of Telomere repeats-binding bouquet protein1
Isoform 2 of Telomere repeats-binding bouquet protein1
Mediator of RNA polymerase II transcriptional subunit 16 OS
Interleukin-1 receptor-associated kinase 4OS |
-
-
-
-
-
-
-
|
-
-
-
-
-
-
-
|
1
3
4
2
2
3
2 |
2
1
1
1
1
3
1 |
2
3
5
4
5
3
1 |
11
7
8
7
8
20
4 |
246
246
768
250
705
865
453 |
-
-
-
-
-
-
-
|
-
-
-
-
-
-
-
|
Table 3. Details of spot analysis: GnRH treated Groups
|
|
Regulation of protein |
Accession No. |
|
|
|
Proteins |
Unique Proteins |
Pep-tides |
PSMs |
AAs |
Molecular wt. (KDa) |
Cal. pI |
-
|
|
-
|
Protein FAM 117B OS |
-
|
-
|
-
|
-
|
1 |
3 |
584 |
-
|
-
|
-
|
|
-
-
-
|
Collagen alpha-2(XI)chain
Protein FAM83 OS
Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosamine |
-
-
-
|
-
-
-
|
-
-
-
|
-
-
-
|
2
2
2 |
2
2
2 |
2
2
2 |
-
-
-
|
-
-
-
|
-
|
|
-
|
Complement component 1Q subcomponent binding protein, mitochondrial
|
-
|
-
|
-
|
-
|
2 |
2 |
278 |
-
|
-
|
-
|
|
-
-
|
Adrenodoxin, mitochondrial
Coiled-coil domain containing protein 79 OS
|
-
-
|
-
-
|
-
-
|
-
-
|
6
1 |
11
2 |
188
768 |
-
-
|
-
-
|
-
|
|
-
|
Tropomyosin beta chain
|
-
|
-
|
-
|
-
|
9 |
13 |
284 |
-
|
-
|
-
|
|
-
-
|
Tropomyosin alpha-1 chain
Coiled-coil domain containing protein 79 OS
|
-
-
|
-
-
|
-
-
|
-
-
|
3
1 |
5
2 |
284
768 |
-
-
|
-
-
|
-
|
|
-
|
Adrenodoxin, mitochondrial
|
-
|
-
|
-
|
-
|
7 |
12 |
188 |
-
|
-
|
-
|
|
-
-
|
Coiled-coil domain containing protein 79 OS
14-3-3 Protein zeta/delta (Fragments)
|
-
-
|
-
-
|
-
|
-
|
1 |
2 |
768 |
-
-
|
-
-
|
-
|
|
-
-
-
|
Coiled-coil domain containing protein 79 OS
Telomere-associated protein RIF1 OS
Transmembrane protein serine 11FOS |
-
-
-
|
-
-
-
|
-
-
-
|
-
-
-
|
1
1
2 |
2
1
2 |
768
2419
439 |
-
-
-
|
-
-
-
|
-
|
|
-
-
-
|
Coiled-coil domain containing protein 79 OS
Neurobeachin-like protein2 OS
Transmembrane protease serine 11F OS |
-
-
-
|
-
-
-
|
-
-
-
|
-
-
-
|
1
3
1 |
2
4
1 |
768
2742
439 |
-
-
-
|
-
-
-
|
-
|
|
-
-
-
|
Annexin A5
Coiled-coil domain containing protein 79 OS
Inactive ubiquitin carboxyl-terminal hydrolase 53
|
-
-
-
|
-
-
-
|
-
-
-
|
-
-
-
|
3
1
1 |
5
1
1 |
319
768
1069 |
-
-
-
|
-
-
-
|
-
|
|
-
-
-
|
Phosphatidylethanolamine-binding protein 1
Coiled-coil domain containing protein 79 OS
PHD finger protein 10 OS
|
-
-
-
|
-
-
-
|
-
-
-
|
-
-
-
|
4
1
1 |
5
2
1 |
187
768
497 |
-
-
-
|
-
-
-
|
-
|
|
-
-
-
|
Coiled-coil domain containing protein 79
G2/mitotic-specific cyclin-B1 |
-
-
-
|
-
-
-
|
-
-
-
|
-
-
-
|
2
1
1 |
3
1
1 |
135
768
430 |
-
-
-
|
-
-
-
|
-
|
|
-
P67778
-
|
-
Prohibitin OS
Neurobeachin-like protein 2 |
-
-
-
|
-
-
-
|
-
-
-
|
-
-
-
|
1
4
1 |
4
4
4 |
246
272
1073 |
-
-
-
|
-
-
-
|
-
|
|
-
-
-
|
ATP synthase subunit d, mitochondrial
Anionic trypsin-2 OS
Transmembrane protease serine 11 F
|
-
-
-
|
-
-
-
|
-
-
-
|
-
-
-
|
3
1
3 |
3
1
3 |
161
246
439 |
-
-
-
|
-
-
-
|
-
|
|
-
-
|
Coiled-coil domain containing protein 79
Transmembrane protease serine 11F |
-
-
|
-
-
|
-
-
|
-
-
|
1
2 |
2
2 |
768
439 |
-
-
|
-
-
|
-
|
|
-
-
-
|
Superoxide dismutase [Cu-Zn]
Coiled-coil domain containing protein 154 OS
ELAV- LIKE PROTEIN 1 OS |
-
-
-
|
-
-
-
|
-
-
-
|
-
-
-
|
2
2
1 |
3
2
1 |
154
657
326 |
-
-
-
|
-
-
-
|
-
|
|
-
-
|
-
Isoform 2 of Protein homolog OS |
-
-
|
-
-
|
-
-
|
-
-
|
1
2 |
4
3 |
246
328 |
-
-
|
-
-
|
Immunoblotting
Two to three ovaries were pooled and homogenized in suspension buffer (0.01 M Tris pH 7.6, 0.001 M EDTA pH 8.0, 0.1 M NaCl, 1 µg/ml aprotinin, 100 µg/ml PMSF) via sonication to produce 10% (w/v) homogenate. Further, protein extraction and immunoblot was performed as described previously in our study. Protein quantification was done using Lowry’s method. Equal amount of proteins (40 µg) were loaded onto SDS–PAGE (10%) for electrophoresis. Separated proteins were then transferred on PVDF membrane (Millipore, Billerica, MA, USA) overnight at 4°C. Non-specific sites were blocked for 60 min with Tris-buffered saline (TBS; Tris 50 mM (pH 7.5), NaCl 150 mM, 0.02% Tween 20) containing 5% fat-free dry milk and thereafter incubated with 14-3-3 zeta/delta antibody at dilution of 1:4000; and SOD1 antibody at a dilution of 1:3000; Prohibitin antibody, PCNA and BCl2 antibody at the dilutions 1:1500; PARP-1 and Caspase-3 at the dilution of 1:1000 (Table 1) for 1 h at room temperature followed by washing for 30 min. Immunodetection was performed with secondary antibody conjugated with horseradish peroxidase (1:4000 v/v) followed by three washings with PBS (0.2 M PBS, pH = 7.4) for 10 min each. Finally, signal was detected with enhanced chemiluminescence (ECL) detection system (Bio-Rad, USA). Equal loading was confirmed either by Ponceau S staining or by β-actin (Sigma). Experiments were repeated three times with the same result.
Densitometry
X-ray films were later scanned and then quantified by densitometry (Image J vs. 1.36, NIH, USA). Quality of loading and transfer was assessed with Ponceau S staining and/or β-actin. All immunoblots were normalized to β-actin. The data are presented as the ± S.E.M. of three blots of each peptide.
Progesterone assay
We have also measured progesterone because progesterone in proestrus was necessary for the induction of apoptosis in the regressing corpora lutea [33]. Hormone assay for progesterone was performed using LDN (Labor Diagnostika Nord GmbH & Co. KG) ELISA KIT. 25µl of test sample was taken and processed as per manufacturer’s instruction.
Statistical analysis
The data were analyzed by one-way analysis of variance (ANOVA) followed by Duncan’s multiple range post hoc test by using SPSS software 16 for Windows (SPSS Inc., Chicago, IL, USA) to compare the data from different groups. All data are expressed as means ± SEM. The data were considered significant if p<0.05.
Results
Proteomics analysis in the ovary of mice treated in vitro with either GnRH or GnIH
Two-dimensional (2D) gel electrophoresis followed by PDQuest analysis was undertaken to evaluate differentially expressed proteins of the ovary of mice treated in vitro with either GnRH or GnIH as compared to the protein profiles of the control ovary. The control ovary showed 681 spots after colloidal CBBG-250 staining. There were 25 differentially expressed protein spots, out of which we have selected 18 differentially protein spots from each treatment group using non-targeted apporoach for LC-MS/MS analysis. (Figures 1-3)
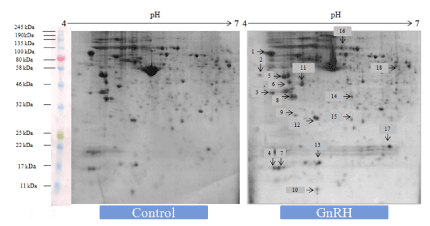
Figure 1. Representative 2-DE gel image showed effects of in vitro treatment of the GnIH (100 ng/ml) on the protein expression profile of the proestrus mice ovary with control ovary. There was a significant variation in the proteomic profile. 15 out of 18 proteins were significantly down-regulated where as other three proteins showed significant up-regulation with control ovary (devoid of neuropeptide). The arrows represent significantly upregulated or downregulated protein spots
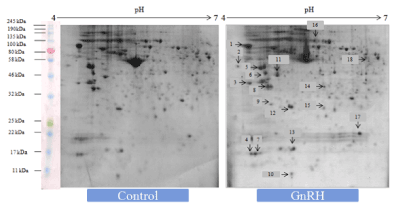
Figure 2. Representative gel image showed effects of in vitro treatment of the GnRH (100 ng/ml) on the protein expression profile of proestrus mice ovary with control. Out of 18 identified GnRH-induced proteins, 9 showed upregulation while 9 proteins were significantly down-regulated. The arrows represent significantly upregulated or downregulated protein spots
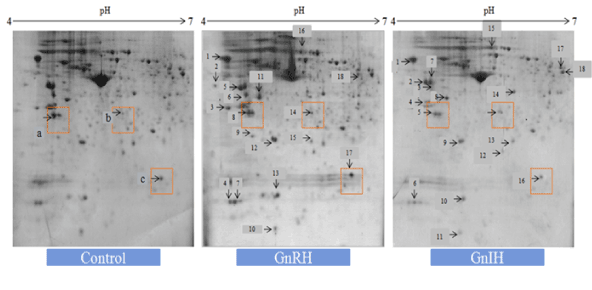
Figure 3. The figure represents effects of in vitro treatment of the GnIH/GnRH on the protein expression profile of the proestrus mice ovary. There was significant variation (at least 2 fold change) either upregulation or downregulation as determined by Student t-test (p<0.05) observed in the proteomic profile to control ovary (devoid of neuropeptide). There were three zones highlighted in red named as a, b and c used for representation of 14.3.3, prohibitin and SOD-1 respectively
Identification of differentially expressed protein spots from the ovary of mice treated with either GnRH or GnIH by mass spectrometry (LC-MS/MS)
Each of the 18 differentially expressed (selected) protein spots separated from the ovaries of mice treated in vitro with either GnRH or GnIH in comparison to protein profile the control ovary was subjected to LC–MS/MS analysis followed by database search against the mouse, Swiss-Prot databases using Proteome Discoverer 3.1 software (Thermo). Each protein spot produced one or two significant hits as determined by false discovery rates of 1.0 and 5.0% using a reverse database. Protein identification was considered confident when 20% sequence coverage was found or the first hit from the database search. The proteins identified are shown in Table 2.
Validation of proteomic study by Western blot analysis of differentially expressed proteins
In order to validate/verify the data from 2 D proteomic analysis, three proteins, 14-3-3, prohibitin and SOD1, were selected. These selected proteins have undergone significant variation (either up- or down-regulated) in the ovary treated in vitro with either GnRH or GnIH as compared with the protein profile of control ovary. Western blot followed by densitometric analysis of the proteins 14-3-3, prohibitin and SOD 1 in the ovary of mice showed a single immunoreactive band at ~ 30, 30 and 18 kDa respectively and results are shown in Figures 4-6.
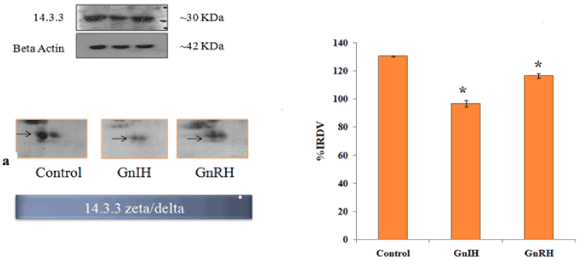
Figure 4. The figure represents effects of in vitro treatment of the GnIH/GnRH on the protein expression profile of proestrus mice ovary. Here the gel images of 14-3-3 were excised and zoom out to see the variation from original gel showed in Figure 3 and designated as a. The protein spots were validated with Western blotting of 14-3-3 protein. The arrow indicates the protein spot of 14-3-3. Values are represented as the mean ± SEM. Value (*) is significantly different (p <0.05) in the treatment groups. IRDV = integrated relative density value
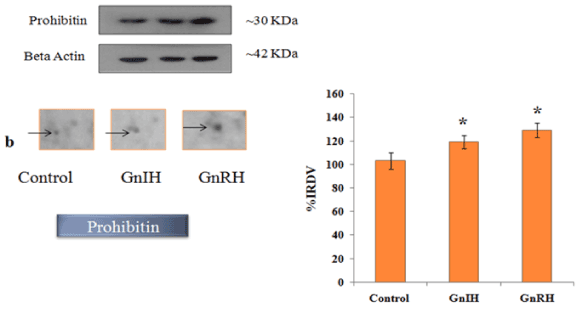
Figure 5. The figure represents effects of in vitro treatment of the GnIH/GnRH on the protein expression profile of proestrus mice ovary. The gel images of prohibitin were excised and zoom out to see the variation from original gel showed in Figure 3 and designated as b. The protein spots were validated with Western blotting of prohibitin protein. The arrow indicates the protein spot of prohibitin. Values are represented as the mean ± SEM. Value (*) is significantly different (p <0.05) in the treatment groups. IRDV = integrated relative density value
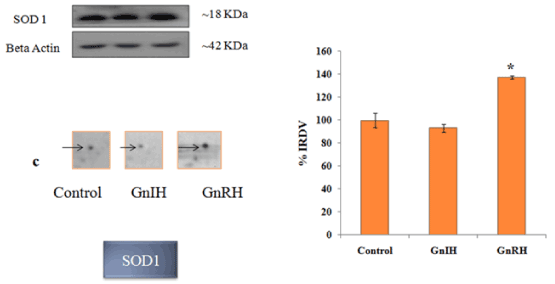
Figure 6. The figure represents effects of in vitro treatment of the GnIH/GnRH on the protein expression profile of proestrus mice ovary. The gel images of SOD1 were excised and zoom out to see the variation from original gel showed in Figure 3 and designated as c. The protein spots were validated with Western blotting of SOD1 protein. The arrow indicates the protein spot of SOD1. Values are represented as the mean ± SEM. Value (*) is significantly different (p <0.05) in the treatment groups. IRDV = integrated relative density value
Effects of in vitro treatment of GnRH and GnIH on the ovarian synthesis of progesterone
The effects of GnRH and GnIH on ovarian progesterone synthesis in vitro are shown in Figure 7. Both the treatments caused a significant decrease in the progesterone synthesis as compared with the control. It was observed that treatment of GnIH showed a more decrease in the progesterone synthesis as compared to GnRH treated group.
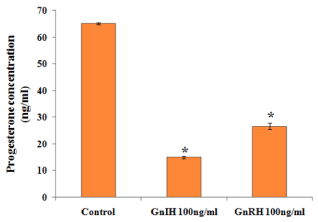
Figure 7. Representative graph showed effects of in vitro treatment of the GnIH/GnRH on the progesterone release from the proestrus ovary and compared with the control group. Values are represented as the mean ± SEM. Value (*) is significantly lower (p <0.05) in the treated group to control
Effects of in vitro treatment of GnRH and GnIH on ovarian marker for follicular growth and Atresia
To investigate the role of GnRH/GnIH, changes in the expression of follicular growth markers (BCl2 and PCNA) and markers for follicular atresia (caspase-3 and PARP-1) were examined after the ovary treated in vitro with these neuropeptide (GnRH/GnIH). The results showed a significant increase in the caspase-3 expression in the ovary treated with either GnRH or GnIH as compared to the control ovary. The in vitro treatment with GnIH caused only a marginal increase in the expression of both whole and cleaved PARP-1, but the treatment with GnRH caused a significant increase in whole PARP-1 compared to the control (Figure 8). The expression of BCl2 and PCNA increased significantly in GnRH treated ovary compared to the control, while the BCl2 expression decreased, but the PCNA expression increased significantly in the GnIH treated ovary as compared to control (Figure 9).
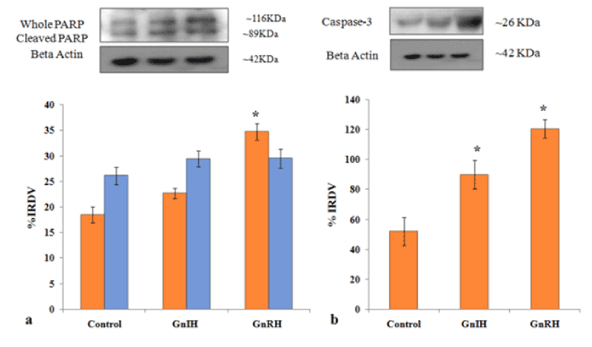
Figure 8. Representative figure demonstrate the effects of in vitro treatment of GnIH/GnRH on the ovarian expression of apoptotic markers (PARP-1 and caspase-3) protein. Values are represented as the mean ± SEM. (a) Immunoblot analysis showed the increased expressions of PARP-1 protein in the ovary of treated mice with control. (b) Immunoblot analysis showed the increased expressions of caspase-3 protein in the ovary of treated mice with control. Values are represented as the mean ± SEM. IRDV = integrated relative density value
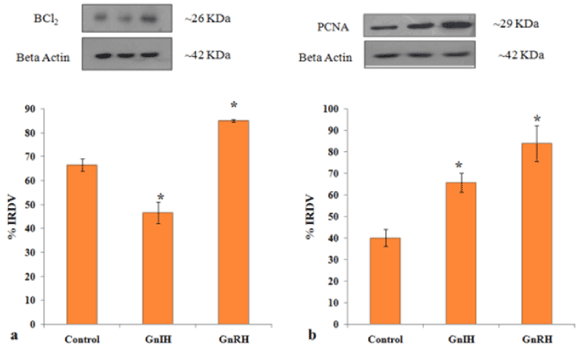
Figure 9. Representative figure demonstrate the effects of in vitro treatment of GnIH/GnRH on the ovarian expression of cell survival marker (BCl2) and cell proliferative marker (PCNA). Values are represented as the mean ± SEM. (a) Immunoblot analysis showed increase expressions of BCl2 protein in the ovary of GnRH treated mice with control whereas there was a decreased expression of BCl2 in the ovary of the GnIH treated group. (b) Immunoblot analysis showed the increased expressions of PCNA protein in the ovary of treated group compared to control. Values are represented as the mean ± SEM. Value (*) is significantly different (p< 0.05) in the treatment group to the control group. IRDV = integrated relative density value
Effects of in vitro treatment of GnRH and GnIH on the ovarian expression of 14-3-3, prohibitin and SOD-1 proteins and their correlation with the markers for follicular growth (PCNA and BCl2) and atresia (PARP-1 and caspase-3) and progesterone synthesis
To achieve this, the ovaries were treated in vitro with either GnRH or GnIH and examined changes in expression of 14-3-3 protein, prohibitin, SOD-1, PCNA, BCl2, PARP-1 and caspase-3 proteins in comparison to the expression of these proteins in the control ovary. The changes found in 14-3-3, prohibitin and SOD-1 were correlated with changes in expression of PCNA, BCl2, PARP-1 and caspase-3 proteins and with changes in the levels of progesterone. The results are given in Table 4. The in vitro treatment with GnIH caused only a marginal increase in the expression of both whole and cleaved PARP-1, but the treatment with GnRH caused a significant increase in whole PARP-1 compared to the control. Changes in the expression of BCl2 and PCNA increased significantly in GnRH treated ovary compared to control, while the BCl2 expression decreased, but the PCNA expression increased significantly in the GnIH treated ovary compared to the control (Table 4).
Table 4. Correlation study among changes in 14.3.3 zeta/delta, Prohibitin, SOD 1, apoptotic markers (whole PARP-1, cleaved PARP-1 and Caspase-3), cell proliferation (PCNA) and cell survival marker (BCl2) by the ovarian cells in the proestrus mice ovary treated in vitro with different peptides GnIH and GnRH-Ag with dose of 100 ng/ml each and control (without any peptide)
|
Prohibitin
|
SOD 1
|
PARP-1 w
|
PARP-1 c
|
Caspase-3
|
BCl2
|
PCNA
|
14.3.3 zeta/delta
|
-0.53115
|
# |
# |
-0.80103
|
# |
0.603508
|
# |
Prohibitin |
NA |
0.696654
|
0.917684
|
0.932667 |
0.997538
|
# |
0.999279
|
SOD 1 |
|
NA |
0.924342
|
# |
0.745245
|
0.918004
|
0.723394
|
PARP-1 w
|
|
|
NA |
0.712568
|
0.943285
|
0.697234
|
0.932109
|
PARP-1 c
|
|
|
|
NA |
0.905076
|
# |
0.918296
|
Caspase-3
|
|
|
|
|
NA |
# |
0.999482
|
BCl2
|
|
|
|
|
|
NA |
#
|
PCNA
|
|
|
|
|
|
|
NA
|
Note: # represents not significant; Data is significant upto p<0.05
Discussion
The presence of not only mature peptides mRNAs of GnRH and GnIH but also their receptors in the ovary of several mammalian species clearly suggest the local synthesis and action of these neuropeptides in the ovary [9,19,34]. The intra-ovarian productions of GnRH and GnIH have been reinforced by direct effects of these neuropeptides on many ovarian functions such as steroidogenesis, follicular growth and atresia and luteolysis. However, the mechanisms by which GnRH and GnIH regulates these functions in the ovary have not so far been evaluated in detail. To our knowledge, this is the first study that investigated changes in the expression of various functional proteins induced by GnRH and GnIH in the ovary of mice. The present study was undertaken to collect information about functional proteins that may be potentially important for mediating effects of GnRH/GnIH in different ovarian activities in the mice.
The 18 spots from each of the protein profiles of GnRH or GnIH-treated ovaries were randomly selected, which showed most significant differential expressions in comparison with the untreated control ovary. These selected protein spots were subsequently analyzed by LC-MS/MS. Comparison of changes in protein expressions between ovaries treated with GnRH or GnIH may allow for identification of proteins involved in specific regulatory activities by which these neuropeptides modulate different ovarian functions. The identified proteins were either up-regulated or down-regulated and were presumed to be involved in a variety of ovarian processes including cell-cycle regulation, cell proliferation, apoptosis, signaling mechanism, regulation of reactive oxidative stress, steroidogenesis, oocyte maturation and regulation of ovarian cancer and tumor development. The results further showed that many of these proteins were expressed commonly in both the ovaries treated either with GnRH or GnIH. These observations suggest that both GnRH and/or GnIH act through some common cascade of signaling pathways and may also be modulating some common biological effects. The common proteins expressed prominently in both the ovaries treated either with GnRH or GnIH includes annexin-A5, 14-3-3 protein, galectin-1, tropomyosin alpha-1 chain, tropomyosin beta-1 chain, adrenodoxin, and phosphatidylethanolamine-binding protein.
The results of Western blot and proteomic findings showed that the expression of 14-3-3 significantly down-regulated, whereas the expression of prohibitin significantly up-regulated in the ovaries treated with either GnRH or GnIH as compared with the control. In contrast, the protein SOD-1 showed a significant up-regulation in GnRH treated ovary but showed no significant variation in GnIH treated ovary when compared with the expression of SOD-1 in the control ovary. Changes in the rate of expression of 14-3-3 protein, prohibitin and SOD-1 proteins in ovaries treated with GnRH or GnIH using western blot analysis were found correlated significantly with changes in the rate of expression of these proteins in proteomic analysis. Thus, the results of Western blot analysis verified the proteomic data.
This study further evaluated the GnRH or GnIH-induced changes in the expression of proteins 14-3-3 protein, prohibitin and SOD-1 and their correlation with different ovarian activities in mice. The results of this study showed a significant correlation between changes in the ovarian concentration of 14-3-3 protein with those in the concentration of cell survival marker (BCl2) but showed inverse correlation with the apoptotic marker (PARP-1) in the ovaries treated with GnRH or GnIH as compared to the control. The role of GnRH in regulating apoptosis in granulosa cells has earlier been well demonstrated [16]. The treatment with GnRH in vivo increases DNA fragmentation, a hallmark of apoptosis, in granulosa cells of preantral and antral follicles [19] and in the corpus luteum in rat [35]. Similar to GnRH, GnIH is also an important intra-ovarian factor causing follicular apoptosis [22]. Based on these findings, it may be hypothesized that the decrease in the expression of 14-3-3 protein induced by GnRH and/or GnIH in the ovary may be responsible for suppressing survival marker and increasing apoptotic activity. These findings suggest that GnRH or GnIH-induced increased expression of caspase-3, PARP-1 and PCNA proteins may be responsible for increasing apoptotic activity as well as cell proliferation in the ovary. Prohibitin is a highly conserved mitochondrial protein that is associated with apoptosis in luteal cells (luteolysis) [36]. Previous studies also provided evidence that prohibitin may have an anti-apoptotic role in undifferentiated granulosa cells which consequently resulted in an increased rate of cell proliferation [36]. As follicles develop toward early or large antral stages, more intense prohibitin expression was observed in the ovary. This suggests a possible role of GnRH in granulosa cells proliferation. Prohibitin has anti-proliferative action on tumor, so it has tumor suppressive property [37]. These findings suggest that GnRH induced increased SOD1 level may be responsible for increased cell survival and proliferation markers in the ovary. This observation is in agreement with the earlier findings that SOD1 is required for the normal development of antral follicles and corpus luteum [38]. In an earlier study, the SOD1 alone, even in the absence of FSH, was shown to inhibit apoptosis in cultured large antral follicles [39]. In ovary SOD1 are normally expressed in theca cells of the antral follicles [40] and plays an important role in the protection of granulosa cells or theca cells from superoxide radicals generated during the increased steroid synthesis and active follicular development [41]. It has earlier been shown that the insufficient level of SOD1 may cause apoptosis in granulosa cells and follicular atresia [42].
Majority of the GnRH or GnIH-induced differentially expressed ovarian proteins selected and identified in the present proteomic study were shown to be associated with cell proliferation, apoptosis, development and progression of cancer [43]. Most of these proteins, such as dynamin, galectin, AT-rich interactive domain-containing protein, kinesin, interleukin-1 receptor-associated kinase, Src family kinase inhibitors, were down-regulated in the ovary upon the treatment with GnIH and have been previously shown to be associated with development and progression of cancer. The contribution of dynamin-1 in genesis of cancer has recently been demonstrated [44]. It has been shown that dynamin-1 causes cancer cell proliferation and metastasis. GnIH-induced down-regulation of dynamin-1 protein may protect ovary from carcinogenesis. The GnIH-induced galectin-1 is a multivalent carbohydrate binding protein that mediates the malignant cellular activities by cross-linking glycoproteins in the tumor microenvironment [45]. Galectin-1 exerts distinct biological effects in various tissues and involved in cell migration, differentiation, proliferation and apoptosis [46-49]. Further, in vitro study showed a stimulatory effect of galectin-1 in granulosa cells proliferation in pig ovary [50]. Galectin-1 also plays an important role in modulating an inflammatory role in the female reproductive system and may be related to multiple pathological conditions, such as endometriosis [50]. Subsequent study showed the increased expression of galectin-1 in human ovary carcinoma cells, which supports a role of galectin-1 in modulation of the cancer cells [51]. The present study showed another ovarian protein AT-rich interactive domain-containing protein (ARID), which was down-regulated upon the treatment with GnIH and is a putative tumor suppressor in human endometrium. The ARID is frequently mutated and contributes to the pathogenesis of ovarian clear cell carcinoma [52,53]. The present study showed down-regulation of chromosome–associated protein kinesin in the ovary treated with GnIH. The importance of kinesins has become recently evident in tumor development and progression by coordinating mitosis and cytokinesis [54]. The interleukin-1 receptor-associated kinase 1 is partially responsible for IL1-induced up-regulation of the transcription factor NF-kappa B and may play an important role in ovarian cancer [55,56]. IL-1 is known to be critically involved in ovarian carcinogenesis and in other solid tumor [57]. Src family kinase members have also been shown to be important in the development of many solid tumor types [58]. In the present study, the GnIH-induced down-regulation in the expression of Src family kinase inhibitor protein was found in the ovary. This present observation supports the earlier proposal that the Src family kinase inhibitors may be used for the treatment of solid tumors [59]. Our findings indicate that GnIH-induced inhibition of proteins, such as dynamin-1, galectin-1, kinesins, interleukin-1 receptor-associated kinase, Src family kinase inhibitors, and ARID, are mainly involved in protecting carcinogenesis, thus these proteins are promising novel therapeutic targets for ovarian cancer treatment. The present findings are valuable for further investigation to reveal the mechanism of carcinogenesis in human ovary.
The role of GnRH as a negative regulator of proliferation in ovarian surface epithelium and ovarian cancer cells has earlier been demonstrated [60,61]. However, the exact mechanism by which GnRH and GnIH modulate ovarian cancer cells remains to be elucidated. The present study showed some of the ovarian proteins which were up-regulated in response to treatment with GnIH or GnRH and found to be linked to the development of cancer. These ovarian proteins include annexin A5, Homeobox protein-engrailed-2, and complement component 1 Q subcomponent binding protein. The present study showed a significant up-regulation in the expression of annexin A5 in the ovaries treated either with GnRH/GnIH in comparison with the untreated control ovary. This present observation corroborated earlier study where GnRH treatment stimulated the synthesis of annexin A5 in pituitary gonadotropes and ovarian luteal cells [62]. Subsequent studies showed that annexin A5 is expressed in many endocrine glands including pituitary, thyroid, adrenal cortex, ovary and testes [63]. The reported relationship between GnRH and annexin A5 in different cell types suggest a common role of annexin A5 following treatment with GnRH. The present study demonstrated for the first time change in the expression of annexin A5 in the ovary treated with GnIH. Annexin A5 is a calcium phospholipid-binding protein that inhibits PKC and PLC and is connected with cell proliferation [64]. It has been suggested that GnRH induced annexin A5 via PKC may result in the inhibition of cell proliferation in uterus [65]. The presence of GnRH, GnIH and GnRH receptor are well documented in the granulosa cells [9]. It is likely that GnRH/GnIH via annexin A5 by modulating cell proliferation may participate in regulation of follicular growth and atresia [62]. Because earlier study also showed the presence of GnRH and annexin A5 in the CL, GnRH and annexin A5 may be involved in luteolysis as suggested [62]. Thus, annexin A5 (or annexin-V) may be considered as a biomarker for the effect of GnRH/GnIH in the ovary. Shibata et al. [64] reported that annexin-V was involved in antiproliferative mechanism in endometrial cancer cells. It is thus possible that GnRH or GnIH-induced up-regulation of annexin-V through PKC may be involved in antiproliferative effect in ovary. GnRH also activates PKC in the ovarian cancer cells [66]. Previous study also showed that GnRH induced activation of PKC is responsible for anti-proliferation in ovarian cancer [67]. Since annexin A5 inhibits PKC and PLC, it has been suggested that GnRH/GnIH-induced up-regulation of annexin A5 expression may be associated with anti-proliferative mechanism in cancer [65].
The present proteomic data also identified proteins, such as homeobox-engrailed and complement component 1 Q subcomponent binding protein (CIQBP), those expressions were up-regulated significantly in the ovary upon the treatment with GnIH or GnRH and they might implicated either directly or indirectly in ovarian carcinogenesis. This study showed significant up-regulation in the expression of homeobox-engrailed protein in the ovary treated with GnIH compared to the control. Engrailed is a member of the homeobox gene family and encodes a transcription factor that is essential during early development. Although many homeobox genes have been reported to be aberrantly expressed in ovarian cancer, the functional significance of engrailed gene in ovarian tumorigenesis requires further investigation [68]. The role of GnRH as the regulator of proliferation in ovary surface epithelium as well as in ovarian cancer cells has been well documented. The present study showed a significant up-regulation of C1QBP protein in the ovary treated with GnRH as compared to the control ovary. The increased expression of C1QBP was shown to be associated with increase in the progression of tumor. C1QBP was also correlated with the expression of PCNA, a known marker of proliferation. Down-regulation of C1QBP expression significantly decreased cell proliferation and growth in T47D cells [69]. Further studies are required to investigate the potential role of homeobox-engrailed and Complement component 1 Q subcomponent binding protein as a biomarker in ovarian cancer.
This study corroborates earlier findings demonstrating the involvement of GnRH/GnIH in the regulation of ovarian steroidogenesis, although the mechanism of these neuropeptides to affect steroidogenesis was not known. The GnRH/GnIH-induced differentially expressed protein associated with steroid biosynthesis identified in the present proteomic study includes adrenodoxin, SOD1, galectin-1, Interleukin-1 alpha associated kinase. Adrenodoxin is an iron-sulfur protein, found in the mitochondria of steroidogenic tissues, which participates in steroidogenesis as an electron transport intermediate for mitochondrial cytochromes P450, including P450scc, the cholesterol side-chain cleavage enzyme [70]. Because two forms of adrenodoxin phosphorylated and unphosphorylated form were found in the ovary, both forms can bind to cytochrome P450scc. Unphosphorylated adrenodoxin acts as a competitive inhibitor of pregnenolone synthesis. Phosphorylated Adrenodoxin is involved in the biosynthesis of all steroid hormones. The presence of galectin-1 in granulosa cells has been reported [71]. In addition, galectin-1 mediated inhibition of FSH-induced P450SCC and 3β-hydroxysteroid dehydrogenase (3β-HSD) gene transcription has been reported. These findings suggest the involvement of galectin-1 as a negative regulator of steroid synthesis in ovaries [50,72]. Interleukin-1 receptor associated kinase plays an essential role in generating IL-1-induced signal transduction mediates an inhibitory effect on ovarian steroidogenesis by suppressing 3β-HSD activity and may be actively involved in the still enigmatic processes of follicular atresia and luteolysis [55,56]. The present study showed up-regulation of adrenodoxin, SOD1 and galectin-1 in the ovary treated with GnRH, whereas all the protein down-regulated upon the treatment with GnIH. However, the present study showed a significant decline in progesterone synthesis by the ovary treated in vitro with either GnRH or GnIH. The reason for this discrepancy is not clear and requires further investigation. In our earlier studies, the ovary treated in vitro with GnRH or GnIH decreased the progesterone synthesis by directly suppressing the expression of 3β-HSD [22,34].
The present study showed both stimulatory and inhibitory effects of GnRH on the ovarian activities. Out of 18 GnRH induced proteins identified in present proteomic analysis, 9 of them showed significantly up-regulation, whereas other 9 showed significant down-regulation as compared with the level of these proteins in the control ovary. These observations suggest that GnRH regulating ovarian activities both by stimulating as well as by inhibiting expression of proteins. The proteins upregulated with the administration of GnRH includes FAM 117B, adrenodoxin, annexin A5, phosphatidylethanolamine-binding protein, galectin-1, prohibitin, ATP synthase, SOD1 and these proteins are mainly associated with the regulation of ovarian cell proliferation, apoptosis and steroidogenesis. The results of the present study support earlier findings demonstrated the role of GnRH in regulating apoptosis in granulosa cells and luteolysis [16,24]. Interestingly the ovaries treated with GnIH showed inhibitory effects on the expression of proteins. Out of 18 proteins analyzed, 15 of these proteins showed significant down-regulation as compared with the control. The present finding suggests that GnIH mainly acts in the ovary by inhibiting the expression of proteins involved in various ovarian activities. The ovary treated with GnIH also showed up-regulation in the expression of some proteins, such as annexin 5A, homeobox-engrailed and nesprin-2. These proteins are associated with cell signaling, proliferation and carcinogenesis. Kif4 was shown to express in oocyte during meiosis [73]. Thus, GnIH-induced down-regulation of Kif4 protein may suggest the inhibition of oocyte maturation in mice. GnRH induced increased synthesis of phosphatidylethanolamine-binding protein (PE) regulates autophagy, whereas GnIH-induced decreased synthesis of PE may increase the production of ROS and induce cell-death [74]. GnIH-induced decreased the synthesis of N4BP2, which plays a role in DNA repair.
These findings suggest that GnRH acts both by stimulating and inhibiting the expression of protein in the ovary. Interestingly, in GnIH-induced ovary, 15 out of 18 proteins were significantly down-regulated, suggesting that GnIH acts mainly by inhibiting the expression of proteins currently involved in ongoing ovarian activities. Most of the GnRH/GnIH-induced proteins identified were presumed to be involved in varieties of ovarian functions including cell-cycle regulation, cell proliferation, apoptosis, signaling mechanism, regulation of reactive oxidative stress, steroidogenesis, oocyte maturation and regulation of ovarian cancer and tumor development. Many of the GnRH/GnIH-induced differentially expressed proteins identified in the present study have been potentially linked to progression and development of cancer. The results of present study thus suggest that GnRH or GnIH-induced changes in ovarian proteins may affect various physiological processes, such as follicular development, atresia, steroidogenesis and cancer. Some of these proteins eventually may serve as diagnostic biomarkers for the detection of ovarian cancer. However, further studies are needed to confirm these findings.
Acknowledgements
We thank Dr. Marvin Karten, Contraceptive branch, NIH, USA for a generous gift of GnRH-Agonist ([DTrp6, Pro9-NEt] GnRH-Ag). We are extremely thankful to Dr. David C. Pallas (Department of Biochemistry, Winship Cancer Institute, Emory University School of Medicine) for the generous gift of 14-3-3 antibody. We would also like to acknowledge the technical assistance of Jane Chu in the analytical chemistry and protein profiling (ACPP) core at Morehouse School of Medicine. AK acknowledge as Centre of Advance Study in Zoology, Banaras Hindu University and DST-Purse, B.H.U for financial support. Anushree Dave, is thankful to UGC, India for providing research fellowship. The authors declare that there is no conflict of interest that would prejudice the impartiality of the scientific work.
References
- Amoss M, Burgus R, Blackwell R, Vale W, Fellows R, et al. (1971) Purification, amino acid composition and N-terminus of the hypothalamic luteinizing hormone releasing factor (LRF) of ovine origin. Biochem Biophys Res Commun 44: 205-210.
- Schally AV, Arimura A, Kastin AJ, Matsuo H, Baba Y, et al. (1971) Gonadotropin-releasing Hormone: One polypeptide regulates secretion of luteinizing and follicle-stimulating hormones. Science 173: 1036-1038.
- Matsuo H, Baba Y, Nair RM, Arimura A, Schally AV (1971) Structure of the porcine LH- and FSH-releasing hormone I. The proposed amino acid sequence. Biochem Biophys Res Commun 43: 1334-1339.
- Aten RF, Polan ML, Bayless R, Behrman HRJ (1987) A gonadotropin-releasing hormone (GnRH)-like protein in human ovaries: Similarity to the GnRH-like ovarian protein of the rat. Clin Endocrinol Metab 64: 1288-1293.
- Birnbaumer L, Shahabi N, Rivier J, Vale W (1985) Evidence for a physiological role of gonadotropin-releasing hormone (GnRH) or GnRH-like material in the ovary. Endocrinology 116: 1367-1370. [Crossref]
- Jones JJ, Hseueh AJ (1980) Direct inhibitory effect of gonadotropin releasing hormone upon luteal luteinizing hormone receptors and steroidogenesis in hypophysectomized rats. Endocrinology 107: 1930-1936.
- Li WI, Jiao S, Chin PP (1993) Immunoreactive gonadotropin-releasing hormone in porcine reproductive tissues. Peptides 14: 543-549.
- Peng C, Fan NC, Lingier M, Vaananen J, Leung PC (1994) Expression and regulation of gonadotropin-releasing hormone (GnRH) and GnRH receptor messenger ribonucleic acids in human granulosa-luteal cells. Endocrinology 135: 1740-1746.
- Singh P, Krishna A, Sridaran R, Tsutsui K (2011) Immunohistochemical localization of GnRH and RFamide-related peptide-3 in the ovaries of mice during the estrous cycle. J Mol Histol 42: 371-381.
- Singh P, Srivastava RK, Krishna A (2016) Effects of gonadotropin-releasing hormone agonist and antagonist on ovarian activity in a mouse model for polycystic ovary. J Steroid Biochem Mol Biol 163: 35-44.
- Schirman-Hildeshens TD, Bar T, Ben-Aroya, N, Koch Y (2005) Differential gonadotropin releasing hormone (GnRH) and GnRH receptor messenger ribonucleic acid expression patterns in different tissues of the female rat across the estrous cycle. Endocrinology 146: 3401-3408.
- Hillensjo T, LeMaire WJ (1980) Gonadotropin releasing hormone agonists stimulate meiotic maturation of follicle-enclosed rat oocytes in vitro. Nature 287: 145-146.
- Ekholm C, Hillensjo T Isaksson O (1981) Gonadotropin-releasing hormone agonists stimulate oocyte meiosis and ovulation in hypophysectomized rats. Endocrinology 108: 2022-2024.
- Hsueh AJ, Jones PB (1981) Extrapituitary actions of gonadotropin-releasing hormone. Endocr Rev 2: 437-461. [Crossref]
- Vaananen JE, Tong BL, Vaananen CM, Chan IH, Yuen BH, et al. (1997) Interaction of prostaglandin F2alpha and gonadotropin-releasing hormone on progesterone and estradiol production in human granulosa-luteal cells. J Reprod Dev 62: 47-56.
- Billig H, Furuta I, Hsueh AJ (1994) Gonadotropin-releasing hormone directly induces apoptotic cell death in the rat ovary: Biochemical and in situ detection of deoxyribonucleic acid fragmentation in granulosa cells. Endocrinology 134: 245-252.
- Pipuette GN, LaPolt PS, Oikawa M, Hsueh AJ (1991) Regulation of luteinizing hormone receptor messenger ribonucleic acid levels by gonadotropins, growth factors, and gonadotropin-releasing hormone in cultured rat granulosa cells. Endocrinology 128: 2449-2456.
- Tilly JL, Billig H, Kowalski KI, Hsueh AJ (1992) Epidermal growth factor and basic fibroblast growth factor suppress the spontaneous onset of apoptosis in cultured rat ovarian granulosa cells and follicles by a tyrosine kinase-dependent mechanism. Mol Endocrinol 6: 1942-1950.
- Leung PC, Cheng CK, Zhu XM (2003) Multi-factorial role of GnRH-I and GnRH-II in the human ovary. Mol Cell Endocrinol 202: 145-153. [Crossref]
- Tsutsui K, Bentley GE, Bedecarrats GT, Osugi T, Ubuka T, et al. (2010) Review: Gonadotropin-inhibitory hormone (GnIH) and its control of central and peripheral reproductive function. Front Neuroendocrinol 31: 284-295.
- Tsutsui K, Bentley GE, Kriegsfeld LJ, Osugi T, Seong JY, Vaudry H (2010) Review: Discovery and evolutionary history of gonadotrophin-inhibitory hormone and kisspeptin: New key neuropeptides controlling reproduction. J Neuroendocrinol 22: 716-727.
- Singh P, Krishna A, Tsutsui K (2011) Effects of gonadotropin-inhibitory hormone on folliculogenesis and steroidogenesis of cyclic mice. Fertil Steril 95: 1397-1404. [Crossref]
- Tsutsui K, Bentley GE (2008) Gonadotropin-inhibitory hormone (GnIH): Biosynthesis, mode of action and functional significance in birds. Avian Biol Res 1: 177-188.
- Zhao S, Zhu E, Yang C (2010) RFamide-related peptide and messenger ribonucleic acid expression in mammalian testis: Association with the spermatogenic cycle. Endocrinology 151: 617–627.
- Gibson EM, Humber SA, Jain S, Williams WP III, Zhao S, Bentley GE, et al. (2008) Alterations in RFamide-related peptide expression are coordinated with the preovulatory luteinizing hormone surge. Endocrinology 149: 4958-4969.
- Anjum S, Krishna A, Sridaran R, Tustsui, K (2012) Localization of Gonadotropin Releasing Hormone (GnRH), Gonadotropin-Inhibitory Hormone (GnIH), Kisspeptin and gnrh receptor and their possible roles in testicular activities from birth to senescence in mice. J Exp Zool A Ecol Genet 317: 630-644.
- Leung PKC, Cheng CK (2004) GnRH as an autocrine regulator in the human ovary. In: The Ovary 2nd edition, 289-304. Eds Leung PKC and Adashi EY. California, USA: Elsevier Academic press
- Son YL, Ubuk T, Millar RP, Kanasaki H, Tsutsui K (2012) Gonadotropin-inhibitory hormone inhibits GnRH-induced gonadotropin subunit gene transcriptions by inhibiting AC/cAMP/PKA-dependent ERK pathway in LßT2 cells. Endocrinology 153: 2332-2343.
- Byers SL, Wiles MV, Dunn SL, Taft RA (2012) Mouse estrous cycle identification tool and images. PLoS One 7: e35538. [Crossref]
- Xiang H, Wang J, Mao Y, Liu M, Reddy VN, et al. (2002) Human telomerase accelerates growth of lens epithelial cells through regulation of the genes mediating RB/E2F pathway. Oncogene 21: 3784-3791.
- Dave A, Krishna A, Tsutsui K (2017) Direct effects of RFRP-1, a mammalian GnIH ortholog, on ovarian activities of the cyclic mouse. Gen Comp Endocrinol 252: 193-199. [Crossref]
- Singh A, Powell MD, Sridaran R, Krishna A (2015) Effects of seasonal adiposity on ovarian activity of Vespertilinoid bat, Scotophilus heathi: Proteomic Analysis. Mol Cell Endocrinol 399: 219-227.
- Gaytán F, Bellido C, Morales C, Sánchez-Criado JE (1998) Both prolactin and progesterone are necessary for the induction of apoptosis in the regressing corpus luteum of the rat. Biol Reprod 59: 1200-1206.
- Singh P, Krishna A (2010) Effects of GnRH agonist treatment on steroidogenesis and folliculogenesis in the ovary of cyclic mice. J Ovarian Res 3: 26. [Crossref]
- Sridaran R, Hisheh S, Dharamrajan AM (1998) Induction of apoptosis by a gonadotropin-releasing hormone agonist during early pregnancy in the rat. Apoptosis 3: 51-57.
- Thompson WE, Asselin E, Branch A, Stiles JK, Sutovsky P, et al. (2004) Regulation of prohibitin expression during follicular development and atresia in the mammalian ovary. Biol Reprod 71: 282-290.
- Manjeshwar S, Branam DE, Lerner MR, Brackett DJ, Jupe ER (2003) Tumor suppression by the prohibitin gene 3'untranslated region RNA in human breast cancer. Cancer Res 63: 5251-5256. [Crossref]
- Matzuk MM, Dionne L, Guo Q, Kumar TR, Lebovitz RM (1998) Ovarian function in superoxide dismutase 1 and 2 knockout mice. Endocrinology 139: 4008-4011. [Crossref]
- Tilly JL, Tilly KI (1995) Inhibitors of oxidative stress mimic the ability of follicle-stimulating hormone to suppress apoptosis in cultured rat ovarian follicles. Endocrinology 136: 242-252.
- Ishikawa M, Tamate K, Nakata T, Sengoku K, Suzuki K, et al. (1993) Superoxide dismutase/corpus luteum: Immunohistochemical localization of Cu,Zn- and Mn superoxide anion in ovulation/luteal function. ARTA 4: 251-259.
- Sugino N (2005) Reactive oxygen species in ovarian physiology. Reprod Med Biol 4: 31-44. [Crossref]
- Suzuki T, Sugino N, Fukaya T, Sugiyama S, Uda T, et al. (1999) Superoxide dismutase in normal cycling human ovaries: Immunohistochemical localization and characterization. Fertil Steril 72: 720-726.
- Wang J, Xu K, Wu J, Luo C, Li Y, et al. (2012) The changes of Th17 cells and the related cytokines in the progression of human colorectal cancers. BMC Cancer 12: 418. [Crossref]
- Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, et al. (2010) Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: Report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol 28: 2529-2537.
- Cousin JM, Cloninger MJ (2016) The role of Galectin-1 in cancer progression, and synthetic multivalent systems for the study of Galectin-1. Int J Mol Sci 17. [Crossref]
- Cooper DN, Barondes SH (1999) God must love galectins; he made so many of them. Glycobiology 9: 979-984. [Crossref]
- Goldring K, Jones GE, Thiagarajah R, Watt DJ (2002) The effect of galectin-1 on the differentiation of fibroblasts and myoblasts in vitro. J Cell Sci 115: 355-366. [Crossref]
- Leffler H (2001) Galectins structure and function--a synopsis. Results Probl Cell Differ 33: 57-83. [Crossref]
- Perillo NL, Marcus ME, Baum LG (1998) Galectins: Versatile modulators of cell adhesion, cell proliferation, and cell death. J Mol Med (Berl) 76: 402-412. [Crossref]
- Walzel H, Brock J, Pohland R, Vanselow J, Tomek W, et al. (2004) Effects of galectin-1 on regulation of progesterone production in granulose cells from pig ovaries in vitro. Glycobiology 14: 871-881.
- Van Den Brule F, Califice S, Garnier F, Fernandez PL, Berchuck A, et al. (2003) Galectin-1 accumulation in the ovary carcinoma peritumoral stroma is induced by ovary carcinoma cells and affects both cancer cell proliferation and adhesion to laminin-1 and fibronectin. Lab Invest 83: 377-386.
- Bi R, Shen X, Zhang W, Cheng Y, Feng Z, et al. (2016) Clear cell carcinomas of the ovary: A mono-institutional study of 73 cases in China with an analysis of the prognostic significance of clinicopathological parameters and IMP3 expression. Diagn Pathol 11: 17. [Crossref]
- Jones S, Wang TL, Shih Ie M, Mao TL, Nakayaman K, et al. (2010) Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 30: 228-231.
- Rath O, Kozielski F (2012) Kinesins and cancer. Nat Rev Cancer 12: 527-539. [Crossref]
- Wesche H, Korherr C, Krancht M, Falk W, Resch K, et al. (1997) The Interleukin-1 Receptor Accessory Protein (IL-1RAcP) is essential for IL-1-induced activation of Interleukin-1 Receptor-associated Kinase (IRAK) and Stress-activated Protein Kinases (SAP Kinases) J Biol Chem 272: 7727-7731.
- Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z (2013) Pillars article: MyD88: An adapter that recruits IRAK to the IL-1 receptor complex. Immunity. 1997. 7: 837-847. J Immunol 190: 5-15. [Crossref]
- Sehouli J, Mustea A, Koensgen D, Lichtenegger W (2003) Interleukin-1 receptor antagonist gene polymorphism in epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev 12: 1205-1208.
- Yeatman TJ (2004) A renaissance for SRC. Nat Rev Cancer 4: 470-480. [Crossref]
- Chen G, Shukeir N, Potti A, Sircar K, Aprikian A, et al. (2004) Up-regulation of Wnt-1 and beta-catenin production in patients with advanced metastatic prostate carcinoma: Potential pathogenetic and prognostic implications. Cancer 101: 1345-1356.
- Emons G, Schroder B, Ortmann O (1993) High affinity binding and direct antiproliferative effects of luteinizing hormone-releasing hormone analogs in human endometrial cancer cell lines. J Clin Endocrinol Metab 77: 1458-1464.
- Kang SK, Choi KC, Cheng KW, Nathwani PS, Auersperg N, et al. (2000) Role of gonadotropin-releasing hormone as an autocrine growth factor in human ovarian surface epithelium. Endocrinology 141: 72-80. [Crossref]
- Yao B, Kawaminami M (2008) Stimulation of annexin A5 expression by gonadotropin releasing hormone (GnRH) in the Leydig cells of rats. J Reprod Dev 54: 259-264. [Crossref]
- Kawaminami M (1998) Immunocytochemical localization of annexin 5, a calcium-dependent phospholipid-binding protein, in rat endocrine organs. Cell Tissue Res 292: 85-89.
- Shibata S, Sato H, Ota H (1997) Involvement of annexin V in antiproliferative effects of gonadotropin-releasing hormone agonists on human endometrial cancer cell line. Gynecol Oncol 66: 217-221.
- Yamamoto H, Sato H, Shibata S, Murata M, Fukuda J, et al. (2001) Involvement of annexin V in the antiproliferative effect of GnRH-Ag agonist on cultured human uterine leiomyoma cells. Mol Hum Reprod 7: 169-173.
- Imai A, Ohno T, Furui T, Takahashi K, Matsuda T, et al. (1993) Gonadotropin-releasing hormone stimulates phospholipase C but not protein phosphorylation/dephosphorylation in plasma membrane from human epithelial ovarian cancer. Int J Gynecol Cancer 3: 311-317.
- Kim KY, Choi KC, Auersperg N, Leung PCK (2006) Mechanism of gonadotropin-releasing hormone (GnRH-Ag)-I and -II-induced cell growth inhibition in ovarian cancer cells: Role of the GnRH-Ag-I receptor and protein kinase C pathway? Endocr Relat Cancer 13: 211-220.
- Kwong KY, Bloom GC, Yang I, Boulware D, Coppola D, et al. (2005) Synchronous global assessment of gene and protein expression in colorectal cancer progression. Genomics 86: 142-158.
- Scully OJ, Yu Y, Salim A, Thike AA, Yip GWC, et al. (2015) Complement component 1, q subcomponent binding protein is a marker for proliferation in breast cancer. Experimental Biol Med 240: 846-853.
- Voutilainen R, Picado-Leonard J, DiBlasio AM, Miller WL (1988) Hormonal and developmental regulation of adrenodoxin messenger ribonucleic acid in steroidogenic tissues. J Clin Endocrinol Metab 66: 383-388.
- Lutomski D, Fouillit M, Bourin P, Mellottee D, Denize N, et al. (1997) Externalization and binding of galectin-1 on cell surface of K562 cells upon erythroid differentiation. Glycobiology 7: 1193-1199.
- Ghizzoni L, Mastorakos G, Vottero A, Barreca A, Furlini M, et al. (1997) Corticotropin-Releasing Hormone (CRH) inhibits steroid biosynthesis by cultured human granulosa-lutein cells in a CRH and interleukin-1 receptor-mediated fashion. Endocrinology 138: 4806-4811.
- Camlin NJ, McLaughlin EA (2017) Kif4 is essential for mouse oocyte meiosis. PLoS One 12: e0170650. [Crossref]
- Rockenfeller P, Koska M, Pietrocola F, Minois N, Knittelfelder O, et al. (2015) Phosphatidylethanolamine positively regulates autophagy and longevity. Cell Death Differ 22: 499-508. [Crossref]









