Conformational study of a strained DNA/RNA by dynamic 1H NMR biospectroscopy and computational methods for molecular modelling, simulation and biopectroscopic studies of DNA/RNA of gum cancer cells facilitates a comprehensive and through understanding of the latest developments in NMR biospectroscopy. It contains explains key breakthroughs in the methodologies and techniques for conformational study of a strained DNA/RNA by dynamic 1H NMR biospectroscopy and computational methods for molecular modelling, simulation and biopectroscopic studies of DNA/RNA of gum cancer cells. Topics include qualitative and quantitative analysis, biomedical applications, conformational study of a strained DNA/RNA by dynamic 1H NMR biospectroscopy and computational methods for molecular modelling, simulation and biopectroscopic studies of DNA/RNA of gum cancer cells.
conformational study, strained DNA/RNA, dynamic 1H NMR biospectroscopy, computational methods, molecular modelling, simulation, gum cancer cells
In the present work, we have performed the experimental and theoretical conformational study of a strained DNA/RNA by dynamic 1H NMR biospectroscopy and computational methods for molecular modelling, simulation and biopectroscopic studies of DNA/RNA of gum cancer cells for the first time. The conformational study of a strained DNA/RNA by dynamic 1H NMR biospectroscopy and computational methods for molecular modelling, simulation and biopectroscopic studies of DNA/RNA of gum cancer cells have been calculated by using ab initio HF and DFT (B3LYP) methods with 6–311++G (d, p) basis set. The conformational study of a strained DNA/RNA by dynamic 1H NMR biospectroscopy and computational methods for molecular modelling, simulation and biopectroscopic studies of DNA/RNA of gum cancer cells were calculated and scaled values are compared with the recorded NMR spectra of the compound. The observed and the calculated frequencies are found to be in good agreement. Furthermore, the thermodynamic and total dipole moment properties of the compound have been calculated in order to get insight into molecular structure of the compound. These computations are carried out with the main aim that the results will be of assistance in the quest of the experimental and theoretical evidence for the title molecule in biological activity and coordination chemistry [1-10].
The NMR spectra of DNA/RNA in gum cancer cells have been recorded and analyzed. The equilibrium geometry, bonding features, and harmonic vibrational frequencies have been investigated by ab initio and density functional theory (DFT) methods. The assignments of the vibrational spectra have been carried out. The computed optimized geometric bond lengths and bond angles show good agreement with experimental data of the title compound. The calculated HOMO and LUMO energies indicate that charge transfer occurs within the molecule. Stability of the molecule due to conjugative interactions arising from charge delocalization has been analyzed using natural bond orbital (NBO) analysis. The results show that the electron density (ED) in the and antibonding orbital and second–order delocalization energies confirm the occurrence of intramolecular charge transfer (ICT). The calculated results were applied to simulate NMR spectra BT which show good agreement with recorded spectra in conformational study of a strained DNA/RNA by dynamic 1H NMR biospectroscopy and computational methods for molecular modelling, simulation and biopectroscopic studies of DNA/RNA of gum cancer cells (Figure 1).
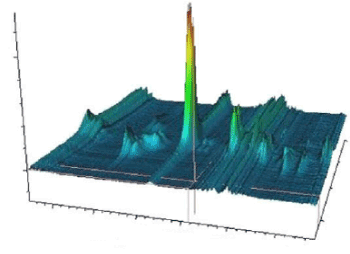
Figure 1. Simulation of strained DNA/RNA in gum cancer cells by dynamic 1H NMR biospectroscopy and computational methods
NMR biomolecular spectroscopic methods, in particular, experimental NMR biospectroscopy, have been successfully employed for structural investigation of complex molecular compounds. These techniques are especially effective when used in combination with direct methods of structural analysis in hydrogen bond investigations for conformational study of a strained DNA/RNA by dynamic 1H NMR biospectroscopy and computational methods for molecular modelling, simulation and biopectroscopic studies of DNA/RNA of gum cancer cells. The aim of the present work is theoretical and experimental biospectroscopic investigation of BT molecular structure to gain insight into the structure and physical properties of the molecular structure for conformational study of a strained DNA/RNA by dynamic 1H NMR biospectroscopy and computational methods for molecular modelling, simulation and biopectroscopic studies of DNA/RNA of gum cancer cells. The NMR spectra were simulated and compared with experimental results. Ab initio and DFT calculations have been performed to support the wave number assignments.
This study was supported by the Cancer Research Institute (CRI) Project of Scientific Instrument and Equipment Development, the National Natural Science Foundation of the United Sates, the International Joint BioSpectroscopy Core Research Laboratory Program supported by the California South University (CSU), and the Key project supported by the American International Standards Institute (AISI), Irvine, California, USA.
- Heidari, C. Brown, “Study of Composition and Morphology of Cadmium Oxide (CdO) Nanoparticles for Eliminating Cancer Cells”, J Nanomed Res., Volume 2, Issue 5, 20 Pages, 2015.
- A. Heidari, C. Brown, “Study of Surface Morphological, Phytochemical and Structural Characteristics of Rhodium (III) Oxide (Rh2O3) Nanoparticles”, International Journal of Pharmacology, Phytochemistry and Ethnomedicine, Volume 1, Issue 1, Pages 15–19, 2015.
- A. Heidari, “An Experimental Biospectroscopic Study on Seminal Plasma in Determination of Semen Quality for Evaluation of Male Infertility”, Int J Adv Technol 7: e007, 2016.
- A. Heidari, “Extraction and Preconcentration of N–Tolyl–Sulfonyl–Phosphoramid–Saeure–Dichlorid as an Anti–Cancer Drug from Plants: A Pharmacognosy Study”, J Pharmacogn Nat Prod 2: e103, 2016.
- A. Heidari, “A Thermodynamic Study on Hydration and Dehydration of DNA and RNA−Amphiphile Complexes”, J Bioeng Biomed Sci S: 006, 2016.
- A. Heidari, “Computational Studies on Molecular Structures and Carbonyl and Ketene Groups’ Effects of Singlet and Triplet Energies of Azidoketene O=C=CH–NNN and Isocyanatoketene O=C=CH–N=C=O”, J Appl Computat Math 5: e142, 2016.
- A. Heidari, “Study of Irradiations to Enhance the Induces the Dissociation of Hydrogen Bonds between Peptide Chains and Transition from Helix Structure to Random Coil Structure Using ATR–FTIR, Raman and 1HNMR Spectroscopies”, J Biomol Res Ther 5: e146, 2016.
- A. Heidari, “Future Prospects of Point Fluorescence Spectroscopy, Fluorescence Imaging and Fluorescence Endoscopy in Photodynamic Therapy (PDT) for Cancer Cells”, J Bioanal Biomed 8: e135, 2016.
- A. Heidari, “A Bio–Spectroscopic Study of DNA Density and Color Role as Determining Factor for Absorbed Irradiation in Cancer Cells”, Adv Cancer Prev 1: e102, 2016.
- A. Heidari, “Manufacturing Process of Solar Cells Using Cadmium Oxide (CdO) and Rhodium (III) Oxide (Rh2O3) Nanoparticles”, J Biotechnol Biomater 6: e125, 2016.

