Human coronaviruses (HCoVs) cause respiratory diseases infecting the upper and/or lower respiratory tract. The six human coronaviruses so far identified are HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU-1, SARS-CoV, and MERS-CoV. Four of these coronaviruses (HCoV-229E, HCoV-OC43, HCoV- NL63, and HCoV-HKU-1) are known as circulating common coronavirus found continuously in the human population causing mostly common cold, with few cases of severe diseases. In late December 2019, a novel human coronavirus, now called SARS-CoV-2, was identified during an outbreak in Wuhan, China. The disease spectrum caused by this virus is now called COVID-19 (Coronavirus Infectious disease 2019). This novel coronavirus has spread globally resulting in a world-wide pandemic that continues to rage as of now. SARS-CoV-2 has a high case morbidity and mortality rate and is high risk to the elderly populations, immune-compromised populations, and to those who have other critical issues like heart disease, diabetes, etc.
In this review, we summarize the latest information of the epidemiology, pathogenesis, and clinical aspects of SARS-CoV-2, and discuss the current scientific and therapeutic advancements for clinical treatment of this pandemic novel coronavirus.
SARS, MERS, covid-19, therapeutics, sars-cov-2, antiviral
In November 2002, a novel cornonavirus, SARS-CoV, was first detected in China, while the larger outbreak was not confirmed until 2003. This virus caused pneumonia and other respiratory complications, referred to as Severe Acute Respiratory Syndrome (SARS). This epidemic spread to 29 countries, and infected about 9000 people with a more than 10% mortality rate [1-5]. SARS was identified as a human coronavirus, adding to four known coronaviruses that primarily cause common cold in humans (HCoV-229E, HCoV-HKU1, HCoV-NL63, and HCoV-OC43) [6-8]. In 2012, MERS-CoV (Middle East Respiratory Syndrome coronavirus) was first identified in Saudi-Arabia, isolated from a human with pneumonia [9]. As of March 12, 2019, a total of 2,494 cases with MERS were found in 27 countries with a mortality rate of more than 30% [10,11]. In December 2019, a new severe acute respiratory syndrome (SARS)-associated coronavirus was identified during an outbreak in Wuhan, China, originally called 2019-nCoV [12]. This new virus primarily causes pneumonia, among other syndromes, and the spectrum of diseases caused by it is called COVID-19 (Coronavirus Infectious Disease 2019). This virus was later named SARS-CoV-2. In March 2020, WHO declared COVID-19 as a global pandemic disease [13]. As of August 10, 2020, 89,270 people in China had been infected with SARS-CoV-2 with more than 4,693 fatalities. In the USA, there have been over 4.9 million confirmed cases with 160K deaths, and worldwide there have been more than 19.7M cases with 728K fatalities [14]. In this review, we will summarize the current knowledge of human coronavirus infection, with emphasis on therapeutic strategies.
A: Human Coronavirus and its Different Types:
Coronaviruses (CoVs) are single-stranded positive-sense RNA viruses whose genome (>27kb) is encapsulated within a lipid membrane envelope carrying spike protein [15]. This envelope is studded with glycoprotein spikes that give coronaviruses their crown-like appearance [16,17]. Table 1 displays the sources of Coronavirus and their classifications.
Table-1: Human Coronavirus and its Different Types [21) |
Cartoon of Human Coronavirus |
|
Hemagglutinin (HE) |
Lipid Bilayer |
Envelope Glycoprotein (E) |
Membrane Protein (M) |
Nucleoprotein (N)+ RNA |
Spike Protein (S) |
|
Common Human Coronaviruses |
Other Human Coronaviruses
(Severe Disease Causing Pathogens) |
|
HCoV-
229E |
HCoV-NL63 |
HCoV-OC43 |
HCoV-HKU1 |
HCoV-SARS |
HCoV-MERS |
HCoV-SARS-2 |
Zoonosis |
Bats à
Camels (?) à
Human |
Bats à
(?) à
Human |
Rodents à
Bovines à
Human |
Rodents à
(?) à
Human |
Bats à
Palm Civets à
Human |
Bats à
Dromedary Camels à
Human |
Bats à
Pangulins (?) à
Human |
Classifications |
a-Type |
a-Type |
b-Type |
b-Type |
b-Type |
b-Type |
b-Type |
Incubation Period |
2-5 days |
2-5 days |
2-5 days |
2-5 days |
2-11 days |
2-13 days |
3-6 days |
Clinical Symptoms |
Malaise,
Headache,
Sneezing, Nasal discharge,
Sore throat, Fever, and Cough |
Cough, Fever, Hypoxia, Croup, Rhinorrhea, Tachypna |
Malaise,
Headache,
Sneezing, Nasal discharge,
Sore throat, Fever, and Cough |
Fever, Running nose, Cough, Dyspnea |
Fever, Myalgia, Headache, Dry cough, Respiratory Distress, Diarrhea, Dyspnea |
Fever, Myalgia, Headache, Cough,
Sore throat, Respiratory Distress, Diarrhea, Dyspnea, Pneumonia |
Fever, Myalgia, Headache, Dry cough,
Respiratory Distress, Diarrhea, Dyspnea, Pneumonia |
Fatality Rates |
None |
None |
None |
None |
~10% |
~35% |
~4% |
A-1. Zoonosis: SARS-CoV is known to spread from bats to civets and then to humans [18], while MERS-CoV spreads from bats to dromedary camels and then to humans [19]. Genome sequencing of SARS-CoV-2 showed that about 96% overall genome sequence were identical with Bat CoV-RaTG13 [20], suggesting that human-SARS-CoV-2 and Bat coronavirus might share a common ancestor [21,22]. However, protein sequence alignment and phylogenetic analysis showed similar residues of receptors in many species, such as turtles, pangolins and snakes [23].
A-2. Classifications: Coronaviruses are divided into four genera: alpha, beta, gamma, and delta [24]. α- and β-CoV are able to infect mammals, while γ- and δ-CoV tend to infect birds. Among the four common human coronaviruses HCoV-229E and HCoV-NL63 are of alpha-type, while HCoV-OC43 and HCoV-HKU1 are in the group of beta-type coronaviruses. These four coronaviruses cause mild to moderate upper-respiratory tract illnesses, including the common cold [25,26]. SARS-CoV, MERS-CoV, and SARS-CoV-2 are all beta-coronaviruses and cause a severe lower respiratory tract infection with higher rates of morbidity and mortality than the α- human coronaviruses [12].
Based on current epidemiological investigation, the incubation period of SARS-CoV-2 ranges from 1–14 days while most commonly it is 3–7 days, similar to other coronaviruses. However, SARS-CoV-2 is known to be contagious during this latency period prior to symptoms appearance [27,28].
A-3. Transmissibility and Pathogenicity:
Coronaviruses express several structural proteins including the glycosylated spike (S) protein that mediates the host cell invasion via binding to a cellular receptor protein. Cell tropism is defined by the entry cellular receptor employed by the coronavirus, via its S glycoprotein. SARS-CoV, SARS-CoV-2, and H-CoV-NL63 utilize Angiotensin-Converting Enzyme-2 (ACE2) as the cellular receptor, whereas MERS-CoV uses Dipeptidyl-Peptidase-IV (DPP-IV), H-CoV-229E uses Amino-Peptidase N (APN), HCoV-HKU1 and HCoV-OC43 employ Sialic Acids as receptors [28-33]. Additionally, all of the α-CoV S-protein contain a sialic-acid binding domain [34,35].
The coronavirus infection process requires S-protein priming, which is facilitated usually by host serine proteases, whether membrane-associated, or extra-cellular. SARS-CoV-2 appears to use cell-bound transmembrane serine protease 2 (TMPRSS2-11?) [11]? from cleavage of S into S1 and S2 upon binding to ACE2 [28]. A portion of the S proteins on the coronavirus virion, may also be cleaved already prior to export of virion outside the cell by proteases in the Golgi apparatus. The viral S glycoprotein thereby consists of two subunits, S1 and S2 [36]. While the receptor-binding-domain (RBD) of S1 protein determines the virus-host range and cellular tropism, the S2 protein with its two tandem domains, heptad repeats 1 (HR1) and heptad repeats 2 (HR2) mediates virus-cell membrane fusion [37,38]. Following fusion, the viral genome is released into the cytoplasm in a Cathepsin L and pH dependent manner, similar to Influenza or Ebola virus (Figure 1) [39-43]. Virion fusion is followed by endocytosis, and uncoating of virion in the cytosol with release of the genomic RNA and virion proteins. These processes are not well studied.
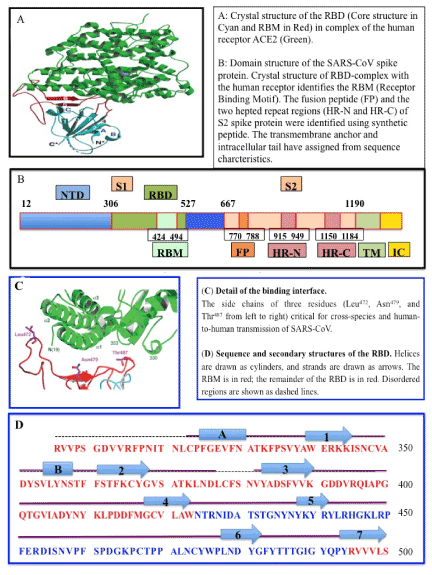
Figure 1. SARS Coronavirus spike receptor-blinding domain (RBD) complexed with ACE2 receptor (A, B), structure of SARS EBD complexed with the receptor (C, D) [108]
Like many other RNA viruses, coronavirus replicates in the cytoplasm of the infected cells. [39-43]. The viral genomic RNA is released into the cytoplasm forming replication-transcription complexes (RTC) in double-membrane vesicles [44-46]. The RTC replicates continuously and synthesize a nested set of subgenomic RNAs [47,48], which encode accessory proteins and structural proteins. The viral genome encodes several nonstructural proteins including RNA-dependent RNA polymerase (RdRp), coronavirus main protease (3CLpro), and papain-like protease (PLpro) [49,50]. The viral RdRp synthesizes a full-length negative-strand RNA template for making more viral genomic RNA. The RdRp also contains a nuclease function with limited proof-reading ability, which is why coronaviruses do not mutate as rapidly as many other RNA viruses such as influenza [51].
Translation of the positive strand viral RNA into two viral polyproteins, pp1a and pp1ab occurs utilizing cellular machinery. This is followed by cleavage into individual effector proteins carried out by viral proteinases, 3CLpro and PLpro [49,50]. PLpro further may deubiquinate certain host cell proteins, including interferon factor 3 and NF-κB, resulting in immune suppression [50,52],
In the endoplasmic reticulum (ER) and Golgi the newly formed genomic +ve RNA, nucleocapsid proteins and envelope glycoproteins assemble and form lipid-coated viral particles [53,54]. Mature coronavirus particles are subsequently transported to the cell surface [46]. Coronaviruses appear to exit cells via cell lysis rather than export/expulsion, although these processes are not well studied. [55].
Cryo-EM structure analysis has revealed that the binding affinity of SARS-CoV-2 S-protein to ACE2 is about 10−20 times higher than that of SARS-CoV S-protein [56,57]. It is speculated that this may contribute to the reported higher transmissibility and contagiousness of SARS-CoV-2 as compared to SARS-CoV [58]. SARS-CoV-2 progeny viruses are released into the luminal side of the human airway, facilitating spread and transmission through droplets and aerosols from coughing and sneezing [10,58,59]. Importantly, sustained human-to-human transmission, along with many exported cases across the globe have led to a world-wide pandemic. However, the disease severity is not necessarily linked to transmission efficiency but may be due to other factors such as age, immunosuppression, or coexisting conditions (such as diabetes or heart disease) [60]. Prior exposure to common cold coronaviruses is thought to provide protection from SARS-CoV-2 resulting in milder disease [61]. Such protection can possibly result from HCoV-NL63 infection, as this virus also uses ACE2 receptor, and its S-protein bears significant similarity with the SARS-CoV-2 S-protein [62].
Zhou et al. have confirmed that the SARS-CoV-2 uses the same cellular entry receptor, ACE2, as SARS-CoV [20]. Yet SARS-CoV-2 is substantially less virulent (~3% mortality) compared to SARS-CoV (~10% mortality) and MERS-CoV (~35% mortality) [10, 63]. HCoV-NL63 also binds to the ACE2 receptor, has become endemic in the human population, but usually causes only a mild upper respiratory tract disease, except in sporadic cases wherein the same clinical syndrome as COVID-19 is reported but with reduced morbidity [23,64]. Thus, the mechanisms of human-to-human transmission and pathogenicity of SARS-CoV-2 reflecting its now-proven pandemic potential are still unknown.
Further, Zhang et al. (2020) recently have found the presence of SARS-CoV-2 in fecal swabs and blood, indicating the possibility of multiple routes of transmission [65]. It is highly transmissible in humans, especially in the elderly and people with underlying diseases [66-69].
A-4. Symptoms of COVID-19: The main clinical symptoms of COVID-19 patients include fever, cough, and fatigue [70]. In addition to the Acute Respiratory Disease Syndrome (ARDS), or Severe Acute Respiratory Syndrome (SARS), the SARS-CoV-2 virus can cause adverse effects in the digestive system, urogenital system, central nervous system, heart, and circulatory system as well [71,72]. Its pathogenicity reflects the broad distribution of ACE2 in human cell lines and tissues, thus causing disease in multiple organs. Additionally, multiple inflammatory syndromes (especially in children, dubbed “MIS-C”), and multi-organ failures can result from the dysregulated attack by the host immune system against the virus directed at clearing out infected cells. People with underlying diseases, like cancer, heart problems, diabetes, and obesity, are susceptible to infection and prone to serious outcomes, which include ARDS, cytokine storm, and death [73, 74].
B. Diagnosis of COVID-19 in Human:
Chest radiography can reveal a typical feature of bronchiolitis with specific features, enabling reliable diagnostics especially in an emergency setting. The most common Chest X-ray pattern was multifocal and peripheral, associated with interstitial and alveolar opacities. In contrast CT exhibited ground glass opacities and was less diagnostic for differentiating the morbidity level of the syndrome [75].
Any unknown pathogen identification is possible using molecular biology tools but may be difficult and time-consuming; however, genome-specific PCR primers can be designed for RT-PCR analysis to specifically detect the virus. FDA has approved several tests on an emergency approval basis for the detection of COVID-19 [76].
Antigen Tests: Detection of one or more SARS-CoV-2 protein antigens in the patient blood. These are quick but not very sensitive nor specific. A positive antigen test means current infection by SARS-CoV-2 or antigen from a different virus that is cross-reactive to the antibodies used in the detection kit. A positive test should be confirmed by a viral RNA test (see below). A negative test means the antigen level in the sample is below detection limit, but does not rule out current infection, so it must be repeated later as the circumstances require.
Nucleic Acid Amplification Tests (Molecular tests): Detection of viral RNA (vRNA) in the human specimen by RT-PCR suggests that the virus might be present. However, if the amount of viral RNA in a particular sample is too low or contaminated with other kind of virus, molecular tests can yield a false negative result. The American Society for Microbiology has developed step-by-step verification procedures for commercial use of COVID-19 diagnostic tests. Both qualitative and quantitative tests are available now.
Antibody Tests (Serology): Generally blood samples are used as a specimen for the presence of antibodies produced in response to SARS-CoV-2 infection, however it does not test for the presence of the virus itself. Therefore, it does not indicate whether a person is currently infected or not, but whether the person was subjected to an infection in the past.
An antibody test can yield a false negative result if the test is conducted too early before the antibodies have developed, or too late, after the antibodies have subsided. It can yield a false positive result if patient antibodies are from a different infection but cross-reactive to SARS-CoV-2 antigen.
The Centers for Disease Control and Prevention (US CDC) has developed a test for SARS-CoV-2 in respiratory and serum samples from infected persons [77]. NIAID (National Institute of Allergy and Infectious Diseases) is also trying to develop additional diagnostic tests for COVID-19. These tests are expected to facilitate preclinical studies and the development of therapeutics [78].
C. Genome Analysis of Coronavirus and its implications to COVID-19 (Figure 1):
All coronaviruses express a set of structural proteins in the order of 5′ - spike (S) - envelope (E) - membrane (M) and nucleocapsid (N) – 3′, as well as some additional nonstructural proteins. The structural gene region also harbors several ORFs that are interspersed along the structural protein coding genes. The location and number of these accessory ORFs vary between the CoV species [79]. The replicase gene is comprised of two overlapping ORFs (open reading frames) that encode up to 16 non-structural proteins required for viral replication and the modulation of any antiviral responses.
Alignment of the HCoV-NL63 and HCoV229E genomes, both alpha-coronaviruses, show the highest sequence similarity, except within the M gene [48]. A careful analysis has shown a large insert of 537 nucleotides are present in the 5′ portion of the S gene of HCoV-NL63 but not in HCoV-229E. A BLAST search, further did not find any similarity between the additional 179–amino acid domain of the S protein of HCoV-NL63 with any other coronavirus sequence deposited in the GeneBank [57,80].
HCoV-NL63 uses ACE2 receptor to bind for invasion and replicates efficiently in monkey kidney cells, as do SARS-CoV, and SARS-CoV-2, though the S proteins of HCoV-NL63 do not match with the two SARS-CoVs sequences [54]. It appears that sequence homology is not the determining factor in receptor binding, and that the RBD quaternary structural features and specific interactions govern binding affinities.
D. Cell Culture Models for COVID-19 Infection:
Different cell culture models are there which can mimic different epithelial tissues [81]. Tracheo-bronchial cells which are the first targets of human respiratory viruses can be cultured in air-liquid interface (ALI). Apical side of the cell layer while exposed to the air, the basolateral side is submerged in the medium. The expression pattern of both primary human bronchial ALI and epithelium in vivo are comparable. Therefore, it appears that these culture methods can be used for virus replication as well as interactions with the host cells [82]. HCoV-HKU1 can propagate in alveolar HAE cultures (Human airway epithelial cell culture), and shows a better tropism for alveolar type II cells [83].
To date, most inhibitors are studied using HCoV-susceptible cell lines. However, an organotypic human airway culture system, since they contain many different cellular sub-populations, may represent an improved system for the testing of new antiviral compounds [84]. Such tissue culture models would require significant efforts for validation of assays and for interpretation of datasets due to heterogeneity of cell populations with variable content.
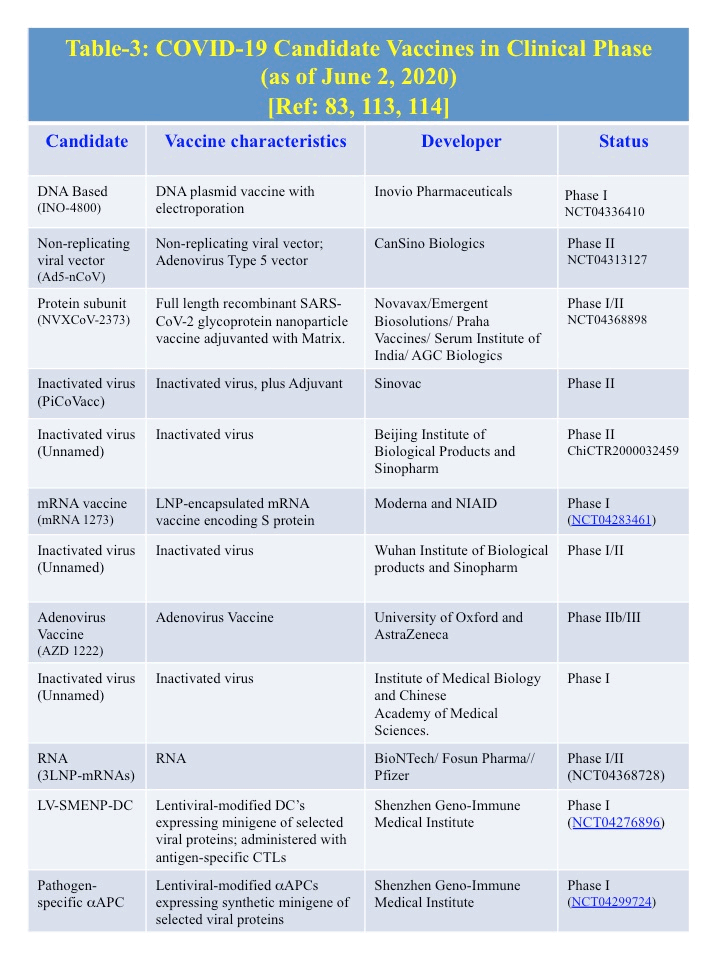
E. Therapeutics (Table 2): As of June 1, 2020, no specific approved treatment for COVID-19 is currently available. Remdesivir and favipravir, both small chemical antivirals that affect replication cycle, have been approved in different countries under emergency use guidelines. With the knowledge of the genome as well as protein structural information from previously discovered six human coronaviruses, several investigators are examining the development of therapeutics. So far 71 treatments and 47 vaccines are in development. Treatments includes various antibodies, antivirals, corticosteroids, immune suppressants, cell-based therapies, and most commonly, re-purposed drugs [85]. Some potential treatments are listed in the Table 2, whose targets are known and also preapproved as a medicine for other disease [23]. Candidate vaccines for COVID-19 that are in clinical phase are listed in the Table 3 [86-88].
Table 2. Some Possible Therapeutics of COVID-19 [23] |
Some Potential Medicines |
Target(s) |
Approved
Medicine for |
Mechanism of action on SARS-CoV-2 |
- Lopinavir
- Rupintrivir
- Small molecule Inhibitor :
(3-1): 4-methyl-N-[(1S)-2-oxo-2 [[(1S,2E)-1-(2-phenylethyl)-3-(phenylsulfonyl)-2-propen-1-yl-amino]-1-(phenylmethyl)ethyl]- 1-piperazinecarboxamide.
(3-2): (αR)-α-[[3-(4-chloro-2-fluorophenyl)-1-oxo-2-propen-1-yl]amino]-N-[(1R)-1-methyl-2-
(2-oxo-3-pyrrolidinyl)ethyl]- benzenepropanamide |
An inhibitor that may disrupt the function of 3CLpro and PLpro, which was tested
against SARS-CoV. |
Lopinavir:
Approved for
HIV infection |
An inhibitor that may disrupt the function of 3CLpro and PLpro, which was tested
against SARS-CoV |
- Remdesivir
- Ribavirin
- Aurine tricarboxylic acid
- Favipiravir (Favilavir)
|
RNA-dependent RNA polymerase (RdRp) for replicating viral genome |
Remdesivir:
Ebola virus infection
Favipiravir:
Viral infections |
An inhibitor of viral RdRp, as tested against SARS-CoV in cell Culture |
- Arbidol
- Camostat mesylate
|
Arbidol:
A viral surface protein (S) for binding to host cell receptor ACE2/.
Camostat mesylate:
Inhibitor of TMPRSS2 |
Arbidol:
Influenza antiviral Drugs
Camostat mesylate:
Antiviral Dug
|
Arbidol:
Inhibits Viral protein binding to ACE2
Camostat mesylate:
Prevents cleavage of the spike protein in SARS-CoV-2, which is necessary for the virus to infect cells. |
Benzopurpurin B |
NSP15 (poly(U)-specific
endo-ribonuclease |
|
Suppress viral infectivity by inhibiting
endo-ribonucleae
NSP15 |
Chloroquine |
Endosome/ACE2 |
Malaria |
Elevates endosomal pH and interferes with ACE2 glycosylation |
Antiviral Antibodies |
S-Protein and other antigens |
|
Neutralizing Antibodies |
Nanoviricides: Venus Fly-Trap |
S-Protein |
|
Multipoint binding sites on Virus particles; and destruction |
E-1. Antiviral Compounds: Inhibition of viral attachment to the host cells and entry should be the prime target of several antiviral compounds [28]. Chloroquine, generally known as an anti-malarial drug, interferes with the terminal glycosylation of ACE2, reducing ACE2 levels, and thus limits virus-receptor binding and thereby entry into the host cell [88]. Furthermore, endosomal acidification which is needed for viral genome release into the host-cell cytoplasm may be inhibited by Chloroquine [88], or its more potent chemical derivative, Hydroxychloroquine [20,23], that have shown promise in testing with SARS-CoV-2 in vitro. Recently, Wang et al. have shown that chloroquine effectively inhibits SARS-CoV-2 in vitro [89], however controversies regarding the efficacy vs. toxicity of these therapeutics remain.
Camostat and Nafamostat, inhibitors of TMPRSS2, have shown effectiveness against coronaviruses entry into the host cells (e.g. MERS-CoV) [28, 90]. Further, endocytic entry of β-coronaviruses can be blocked by Imatinib, an Abelson (Abl) kinase inhibitor [91]. All of these potential therapeutics inhibited the replication of MERS-CoV, SARS-CoV, and HCoV-229E in the low-micromolar range, which is indicative of their potential utility as a broad-spectrum antiviral remedies [83].
E-2. Protease Inhibitors to Block Viral Evasion: The viral evasion needs proteases like 3CLpro and PLpro, which help the translation of essential viral polyproteins (pp1a and pp1ab) and then further proteolysis to form effector proteins [92]. Kaletra, a combination of Lopinavir & Ritonavir, inhibits the activity of 3CLpro and is approved for the treatment of HIV/AIDS [93]. Preliminary clinical trials using Kaletra for the treatment of SARS-CoV-2, however were disappointing [94]. Efforts are still going on to test in combination with other antiviral drugs.
Disulfiram, an FDA-approved alcohol-aversion drug, was found to inhibit papain-like protease in MERS-CoV [95]. Numerous other inhibitors are being examined, with promising results against SARS viruses [83].
E-3. Inhibition of Viral replication: A number of components including the viral RNA-Dependent RNA polymerase (RdRp) and Helicase (nsP13) protein are required for the virus replication [92]. The small molecule 1,2,4-triazole is a derivative that inhibited the viral NTPase/helicase of SARS- and MERS-CoVs with low cytotoxicity [96,97], and could be considered for use against SARS-CoV-2.
Remdesivir (GS-5374), an adenosine analog and RdRp inhibitor, developed to combat other viruses (e.g. Ebola virus) has shown strong activity against SARS-CoV-2 in vitro [68,98,99], but only limited effectiveness in some of its clinical trials [100]. While it shows strong activity in cell culture studies, its clinical activity is limited by its rapid metabolic conversion to nearly inactive form, and its maximum dosage is limited by organ toxicities including liver and kidney. Favipravir, another RdRp inhibitor approved for emergency use in novel Influenza in Japan, was approved in Japan and in China for emergency use for COVID-19. However, it has very limited clinical effectiveness, which is comparable to or less than that of remdesivir.
E-4. Immune therapy: The first line of defense against any pathogens is the innate immune system of the host. The receptor determinant N-acetyl-9-O-acetylneuraminic acid or O-Acetylated Sialic acid Interferon (IFN) System coordinates with various cellular effector proteins and impair virus propagation and transmission. In general, early infection with HCoV-229E, MERS- or SARS-CoVs do not express any innate immunity in the primary target cells of the human airway [101]. This is most likely due to the fact that 5′ terminal of the viral mRNA being capped is indistinguishable from the host cellular mRNAs, and therefore not detectable by cellular immune sensors [89,102-104]. In addition, various other CoV accessory proteins inhibit interferon signaling at different stages of the host-innate-immune response [105,106].
Sarilumab and Tocilizumab, the monoclonal antibodies (mAbs) against the IL-6 receptor, could be possible therapeutics for COVID-19 patients, but may develop a risky cytokine storms [107]. Recent in vitro experimental data find more susceptibility of SARS-CoV-2 to type-1 interferon (e.g. Interferon-β) [108].
A combination of treatments such as steroids, interferon, and the antiviral drug Ribavirin has shown an effective antiviral effect [109-112]. Ribavirin is highly toxic to red blood cells.
E-5. Vaccines: Vaccines against the spike proteins (S) of both SARS-CoVs and MERS-CoVs pathogens are promising in animal models [113,114]. A selection of COVID-19 candidate vaccines that are in clinical phase are shown in Table 3. [115-117].
E-6. Animal Model for Vaccine Development: Traditionally, studies with respiratory viruses use mice and ferrets as an animal model. However, no animal model could represent human infection or disease correctly. Currently, there is no validated animal model for SARS-CoV-2 infection, and therefore there is no animal model for testing efficacy of either therapeutics or vaccines for COVID-19.
Further, adapted cell-cultured viruses may not show the same properties as wild-type viruses. SARS-CoV was able to replicate in different strains of inbred mice but to enhance their clinical signs, human ACE2 has been introduced. Transgenic animal models (mouse) expressing human ACE2 have become available. This provides for emulation of virus entry in human cells, but does not mimic the human response to the virus. These models are not yet validated for clinical pathology to the best of our knowledge. However, cells isolated from these transgenic h-ACE2 animals could be used for infection inhibition studies in vitro [118,119].
Considering the emergence of SARS-CoV-2, along with SARS- and MERS-CoVs, the pandemic potential of coronaviruses is now well established, and animal model development is necessary. Currently, a few animal models for MERS-CoV have been established by introducing the human variant of the viral receptor DDP4 (Dipeptidyl Peptidase 4) in place of the mice variant. However, the method of insertion of human Dipeptidyl Peptidase 4 (hDPP4) negatively affects the degree of pathogenesis [120,121]. Various non-human primates (NHPs) can be naturally infected with both MERS-, and SARs-CoVs, but the disease presentation and pathogenesis differs among the subspecies. Furthermore, NHP models are expensive and come with ethical obstacles [122].
E-7. Novel nanomedicine approaches
U.C. SanDiego engineers coated a polymer using an extract of lung cell membranes and demonstrated that this so called “nanosponge” indeed acted as a decoy for the SARS-CoV-2 virus in vitro [123]. Such an undefined materials cannot be advanced into regulatory frameworks.
Considering the merits and demerits of the above approaches, Nanoviricides, Inc., a Shelton (CT) based biotech company dedicated to antiviral research, has developed a platform technology that goes beyond what antibodies can do for attacking a virus. We have developed a “Venus-Fly-Trap” for virus particles. The antiviral drug platform that we call “nanoviricide®” provides multiple binding sites that mimic the cellular receptor on a polymeric micelle in high density. The binding sites are made up of small chemical virus-binding ligands designed using molecular modeling. Virion binding to the nanoviricide would lead to a multi-point attack on the virus particle, with the metastable polymeric micelle inverting and fusing into the lipid membrane of the virus envelop, thereby destabilizing the virus particle. Antibodies, in contrast, only bind to the virus via two points, requiring multiple antibodies to tag the virus, and further require the patient’s immune system to destroy the virion. In addition antibodies are highly specific and single point mutations can allow viruse escape. In contrast, the ligands are designed to be broad-spectrum, and to mimic the conserved features of the binding of virion to its cognate receptor, thereby minimizing the possibility of viral escape (Table 2). In order to assess how broad-spectrum effect our drug candidates may possess we are testing our compounds now against two different coronaviruses, 229E and NL63 (Work is in Progress). (https://www.marketwatch.com/press-release/nanoviricidesdevelops-highly-effective-broad-spectrum-drug-
candidates-againstcoronaviruses-2020-05-12. NanoViricides Develops Highly Effective Broad-Spectrum Drug Candidates Against Coronaviruses. Published: May 12, 2020).
Since no vaccines are currently available for any of the respiratory coronaviruses (SARS, MERS and SARS-CoV-2), it is necessary to investigate the spread of infections and to follow the protection guidelines. In accordance with other recent reviews on this area [74], we agree that more detailed epidemiological studies are required to determine the pathological effects on other body/organ systems. Finally, a more detailed knowledge of the genomes and proteomes of previously described coronaviruses HCoV-NL63, HCoV-229E, SARS-CoV, MERS-CoV, and their role in the pathogenesis, is needed to better understand infection as well as for improved drug design. An appropriate animal model also needs to be developed that mimics the human clinical pathology. Further, the development of an accurate diagnosis of COVID-19 will shed light on the true incidence of this virus infection.
The authors declare no competing interests.
This work was supported by funding from Nanoviricides, Inc (Shelton, CT, USA). All the authors contributed equally.
- WHO (2003) Cumulative number of reported probable cases of SARS. https://www.who.int/csr/sars/country/2003_07_11/en/.
- Holmes EC, Rambaut A (2004) Viral evolution and the emergence of SARS coronavirus. Philos Trans R Soc Lond B Biol Sci 2359: 1059-1065. [Crossref]
- Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, et. al. (2003) Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361: 1767-1772.
- Lai MM (2003) SARS virus: the beginning of the unraveling of a new coronavirus. J. Biomed Sci 10: 664-675. [Crossref]
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, et. al. (2003) Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348: 1967-1976. [Crossref]
- Pene F, Merlat A, Vabret A, Rozenberg F, Buzyn A, et. al. (2003) Coronavirus 229E-related pneumonia in immune-compromised patients. Clin Infect Dis 37: 929-932.
- Vabret A, Mourez T, Gouarin S, Petitjean J, Freymuth F (2003) An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin Infect Dis 36: 985-989. [Crossref]
- Woo PC, Lau SK, Tsoi HW, Huang Y, Poon RW, et. al. (2005) Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J Infect Dis 192: 1898-1907. [Crossref]
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Eng J Med 367: 1814-1820.
- De Wit E, van Doremalen N, Falzarano D, Munster VJ (2016) SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14: 523-534. [Crossref]
- WHO (2019) Middle East respiratory syndrome coronavirus (MERS-CoV), Nov 2019. www.who.int/emergencies/mers-cov/en
- Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Napoli RD (2020) Features, Evaluation and Treatment Coronavirus (COVID-19) In: StatPearls. Treasure Island (FL): StatPearls Publishing.
- Cucinotta D, Vanelli M (2020) Who declares COVID-19 a pandemic. Acta Biomed 91: 157‐160. [Crossref]
- NIAID (2020) Covid-19 is an emerging, rapidly evolving situation.
- Schoeman D, Fielding BC (2019) Coronavirus envelope protein: current knowledge. Virol J 16: 69-91.
- McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM (1967) Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci USA 57:933. [Crossref]
- Masters PS, Perlman S (2013) Coronaviridae. In: Fields Virology, 6th ed, Knipe DM, Howley PM, Cohen JI, et al. (Eds), Lippincott Williams & Wilkins, aWolters Kluwer business, Philadelphia.
- Wang LF, Shi Z, Zhang S, Field H, Daszak P, et al. (2006) Review of bats and SARS. Emerg Infect Dis 12: 1834‐1840. [Crossref]
- Killerby ME, Biggs HM, Midgley CM, Gerber SI, Watson JT (2020) Middle east respiratory syndrome coronavirus transmission. Emerg Infect Dis 26: 191-198.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270-273.
- Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, et al. (2020) Zoonotic origins of human coronaviruses. Int J Biol Sci 16: 1686‐1697. [Crossref]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579: 265-269.
- Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, et. al. (2020) Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci 6: 315-331. [Crossref]
- Wei X, Li X, Cui J (2020) Evolutionary perspectives on novel coronaviruses identified in pneumonia cases in China. Natl Sci Rev 7: 239-242.
- Cui J, Li F, Shi ZL (2019) Origin and Evolution of Pathogenic Coronaviruses Nat Rev Microbiol 17: 81-192.
- World Health Organization (2020) Coronavirus. https://www.who.int/health-topics/coronavirus
- Jin Y, Yang H, Ji W, Wu W, Chen S, et al. (2020) Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses 12: 372. [Crossref]
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, et. al. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271-280
- Du L, He Y, Zhou Y, Liu S, Zheng B-J, et al. (2009) The spike protein of SARS-CoV- A target for vaccine and therapeutic development. Nat Rev Microbiol 7: 226-236. [Crossref]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, et al. (2020) Cryo-EM structure of the 2019-nCoV Spike in the prefusion conformation. Science 10: eabb2507.
- Bonavia A, Zelus BD, Wentworth DE, Talbot PJ, Holmes KV (2003) Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J Virol 77: 2530-2538.
- Godet M, Grosclaude J, Delmas B, Laude H (1994) Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol 68: 8008-8016. [Crossref]
- Krempl C, Ballesteros ML, Zimmer G, Enjuanes L, Klenk HD (2000) Characterization of the sialic acid binding activity of transmissible gastroenteritis coronavirus by analysis of haemagglutination-deficient mutants. J Gen Virol 81: 489-496.
- Huang X, Dong W, Milewska A, Golda A, Qi Y, Zhu QK, et. al. (2015) Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J. Virol. 89: 7202–7213.
- Butler N, Pewe L, Trandem K. (2006) Perlman, S. Murine encephalitis caused by HCoV-OC43, a human coronavirus with broad species specificity, is partly immune-mediated. Virology 347: 410-421.
- Zhang N, Jiang S, Du L (2014) Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev Vaccines 13:761-774. [Crossref]
- Xia S, Zhu Y, Liu M, Lan Q, Xu W, et al. (2020) Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol 10: 1-10.
- Yu F, Du L, Ojcius DM, Pan C, Jiang S (2020) Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect 22: 74-79. [Crossref]
- Millet JK, Whittaker GR (2015) Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202: 120-134. [Crossref]
- Yang ZY, Huang Y, Ganesh L, Leung K, Kong WP, et. al. (2004) pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol 78: 5642-5650.
- Li F, Li W, Farzan M, Harrison SC (2005) Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309: 1864-1868.
- Menachery VD, Yount BL, Debbink K, Agnihothram S, Gralinski LE, et. al. (2016) A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 22: 446. [Crossref]
- Lundin A, Dijkman R, Bergström T, Kann N, Adamiak B, et. al. (2014) Targeting Membrane-Bound Viral RNA Synthesis Reveals Potent Inhibition of Diverse Coronaviruses Including the Middle East Respiratory Syndrome Virus. PLOS Pathogens 10: e1004166.
- de Wilde AH, Snijder EJ, Kikkert M, van Hemert MJ (2018) Host factors in coronavirus replication. Curr Top Microbiol Immunol 419:1-42.
- Sawicki SG, Sawicki DL (2005) Coronavirus transcription: a perspective. Curr Top Microbiol Immunol 287: 31-55. [Crossref]
- Salanueva IJ, Carrascosa JL, Risco C (1999) Structural maturation of the transmissible gastroenteritis coronavirus. J Virol 73: 7952-7964.
- Hussain S, Pan J, Chen Y, Yang Y, Xu J, et al. (2005) Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J Virol 79: 5288-5295. [Crossref]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, et. al. (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of inter subtype recombination. J. Virol 73: 152-160.
- Gorbalenya AE, Snijder EJ, Ziebuhr J (2000) Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol 81: 853-879.
- Baez-Santos YM, St. John SE, Mesecar AD (2015) The SARS coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res 115: 21-38. [Crossref]
- Mine A, Takeda A, Taniguchi T, Hisaaki Taniguchi, Masanori Kaido, Kazuyuki Mise, et al. (2010) Identification and characterization of the 480-kilodalton template-specific RNA-dependent RNA polymerase complex of red clover necrotic mosaic virus. J Virol. 84: 6070-6081.
- Lee TW, Cherney MM, Huitema C, Liu J, James KE, et al. (2005) Crystal structures of the main peptidase from the SARS coronavirus inhibited by a substrate like aza-peptide epoxide. J Mol Biol 353: 1137-1151. [Crossref]
- Perrier A, Bonnin A, Desmarets L, Danneels A, Goffard A, et al. (2019) The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J Biol Chem 294: 14406-14421. [Crossref]
- Ksiazek TG, Dean Erdman D, Goldsmith CS, Zaki SR, Peret T, et. al. (2003) A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348: 1953-1966.
- Kaye M, Druce J, Tran T, Kostecki R, Chibo D, Morris J, et al. (2006) SARS–associated Coronavirus Replication in Cell Lines. Emerg Infect Dis 12: 128-133.
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. (2020) Cryo-EM structure of the 2019-nCoV Spike in the prefusion conformation. Science 367: 1260-1263.
- Lu R, Zhao X, Li J, Niu P, Yang B, et al.2 (2020) Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet 395: 565-574. [Crossref]
- Tang B, Bragazzi NL, Li Q, Tang S, Xiao Y, et al. (2020) An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov). Infect Dis Model 5: 248-255.
- Song Z, Xu Y, Bao L, Zhang L, Yu P, et. al. (2019) From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses 11: E59. [Crossref]
- CDC (2020) People who are at higher risk: https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-at-higher-risk.html
- Chris Baraniuk (2020) Does the Common Cold Protect You from COVID-19? The scientist: Exploring Life and Inspiring Innovation. https://www.the-scientist.com/
- Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pöhlmann S (2005) Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA 102: 7988-7993.
- Weinger JG, Marro BS, Hosking MP, Lane TE (2013) The chemokine receptor CXCR2 and coronavirus-induced neurologic disease. Virology 435: 110-117.
- van der Hoek L, Pyrc K, Berkhout B (2006) Human coronavirus NL63, a new respiratory virus. FEMS Microbiol Rev 30: 760‐773. [Crossref]
- Zhang W, Du RH, Li B, Zheng XS, Yang XL, et al. (2020) Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 9: 386-389. [Crossref]
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, et al. (2020) Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 1: 1-10.
- Li Q, Guan X, Wu P, Wang X, Zhou L, et al. (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382: 1199-1207.
- Wang D, Hu B, Hu C, Zhu F, Liu X, et al. (2020) Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323: 1061-1069 [Crossref]
- Adalja A, Toner E, Inglesby TV (2020) Priorities for the US Health Community Responding to COVID 19. JAMA 323: 1343-1344.
- Poutanen SM, Low DE, Henry B, Finkelstein S, Rose D, et al. (2003) Identification of severe acute respiratory syndrome in Canada. N Engl J Med 348: 1995-2005.
- Zaim S, Chong JH, Sankaranarayanan V, Harky A (2020) COVID-19 and Multiorgan Response. Curr Probl Cardiol 100618. [Crossref]
- Berger JR (2020) COVID-19 and the nervous system. J Neurovirol 26: 143‐148.
- Huang C, Wang Y, Li X, Ren L, Zhao J, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 395: 497-506.
- Guo Y, Cao Q, Hong Z, Tan YY, Chen S-D, et al. (2020) The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Military Med Res 7: 11-21. [Crossref]
- Ippolito D, Maino C, Pecorelli A, Allegranza P, Cangiotti C, Capodaglio C, et. al. (2020) Chest X-ray features of SARS-CoV-2 in the emergency department: a multicenter experience from northern Italian hospitals: in Respiratory Medicine
- American Society of Microbiology COVID-19TestingFAQs: https://www.asm.org/Articles/2020/April/COVID-19-Testing-FAQs
- CDC: Tests for COVID-19: https://www.cdc.gov/coronavirus/2019-ncov/testing-in-us.html.
- FDA: FDA Working With NCI, NIAID on SARS-CoV-2 Serology Test Validation Effort. https://www.360dx.com/molecular-diagnostics/fda-working-nci-niaid-sars-cov-2-serology-test-validation-effort#.XuelempKj_8
- Chu DK, Peiris JS, Chen H, Guan Y, Poon LL (2008) Genomic characterizations of bat coronaviruses (1A, 1B and HKU8) and evidence for co-infections in Miniopterus bats. J Gen Virol 89: 1282-1287. [Crossref]
- van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJM, et al. (2004) Identification of a new human coronavirus. Nat Med 10: 368‐373.
- Qian Z, Travanty EA, Oko L, Edeen K, Berglund A, et. al. (2013) Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am J Respir Cell Mol Biol 48: 742-748. [Crossref]
- Dominguez SR, Travanty EA, Qian Z, Mason RJ (2013) Human coronavirus HKU1 infection of primary human type II alveolar epithelial cells: cytopathic effects and innate immune response. PLoS One 8: e70129. [Crossref]
- Pillaiyar T, Meenakshisundaram S, Manickam M (2020) Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov Today 25: 668-688.
- Jonsdottir HR, Dijkman R (2016) Coronaviruses and the human airway: a universal system for virus-host interaction studies. Virol J 13: 24.
- Le TT, Andreadakis Z, Kumar A, Román RG, Tollefsen S, et al. (2020) The COVID-19 vaccine development landscape. Nature Reviews (Drug Discovery) 19: 305-306.
- WHO (2020) Draft landscape of COVID-19 candidate vaccines https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- Mullard A (2020) COVID-19 vaccine development pipeline gears up. The Lancet 395: 1751-1752.
- Vincent MJ, Bergeron E, Benjannet S, Erickson BR, Rollin PE, et. al. (2005) Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J 2: 69-79. [Crossref]
- Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, et. al. (2011) Ribose 2'-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12: 137-143.
- Yamamoto M, Matsuyama S, Li X, Takeda M, Kawaguchi Y, et. al. (2016) Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome coronavirus s protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother 60: 6532-6539. [Crossref]
- Sisk JM, Frieman MB, Machamer CE (2018) Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors. J Gen Virol 99: 619-630.
- Fehr AR, Perlman S (1282) Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 1282: 1-23.
- Dong L, Hu S, Gao J (2020) Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov Ther 14: 58-60. [Crossref]
- Cao B, Wang Y, Wen D, Liu W, Wang J, et. al. (2020) A Trial of lopinavir-ritonavir in adults hospitalized with severe COVID-19. N Engl J Med 382: 1787-1799.
- Lin MH, Moses DC, Hsieh CH, Cheng SC, Chen YH, et. al. (2018) Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antiviral Research 150: 155-163.
- Shen L, Niu J, Wang C, Huang B, Wang W, et. al. (2019) High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J Virol 93: e00023-19.
- Adedeji AO, Singh K, Calcaterra NE, DeDiego ML, Enjuanes L, et. al. (2012) Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob Agents Chemother 56: 4718-4728. [Crossref]
- Martinez MA (2020) Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrob Agents Chemother 64: e00399-20
- Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, et. al. (2016) Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 531: 381-385.
- Lerner S (2020) Despite the hype, Gilead’s remdesivir will do nothing to end the coronavirus pandemic.
- Kindler E, Jónsdóttir HR, Muth D, Hamming OJ, Hartmann R, et. al. (2013) Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio 4: e00611-12. [Crossref]
- van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, et al. (2008) SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog 4: e1000054.
- Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, et. al. (2008) SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6: e226. [Crossref]
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, et. al. (2010) 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468: 452-456.
- Siu KL, Yeung ML, Kok KH, Yuen KS, Kew C, et. al. (2014) Middle east respiratory syndrome coronavirus 4a protein is a double-stranded RNA binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol 88: 4866-4876. [Crossref]
- Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, et. al. (2013) Middle east respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 87: 12489-12495.
- Luo P, Liu Y, Qiu L, Liu X, Liu D, et al. (2020) Tocilizumab treatment in COVID-19: A single center experience. J Med Virol 92: 814-818. [Crossref]
- Lokugamage KG, Hage A, Schindewolf C, Rajsbaum R, Menachery VD (2020) SARS-CoV-2 is sensitive to type I interferon pretreatment.
- Al-Tawfiq JA, Momattin H, Dib J, Memish ZA (2014) Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int J Infect Dis 20: 42-46.
- Zumla A, Azhar EI, Arabi Y, Alotaibi B, Rao M, et. al. (2015) Host directed therapies for improving poor treatment outcomes associated with the middle east respiratory syndrome coronavirus infections. Int J Infect Dis 40: 71-14. [Crossref]
- Omrani AS, Saad MM, Baig K, Bahloul A, Abdul-Matin M, et. al. (2014) Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect Dis 14: 1090-1095.
- Shalhoub S, Farahat F, Al-Jiffri A, Simhairi R, Shamma O, et. al. (2015) IFNalpha2a or IFN-beta1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J Antimicrob Chemother 70: 2129-2132. [Crossref]
- Bisht H, Roberts A, Vogel L, Bukreyev A, Collins PL, et. al. (2004) Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci USA 101: 6641-6646.
- Volz A, Kupke A, Song F, Jany S, Fux R, et. al. (2015) Protective efficacy of recombinant modified vaccinia virus Ankara delivering Middle East respiratory syndrome coronavirus spike glycoprotein. J Virol 89: 8651-8656. [Crossref]
- Mullard A (2020) COVID-19 vaccine development pipeline gears up. The Lancet 395: 1751-1752.
- WHO (2020) Draft landscape of COVID-19 candidate vaccines. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- Le TT, Andreadakis Z, Kumar A, Román RR, Tollefsen S, Saville M, et. al. (2020). The COVID-19 vaccine development landscape. Nature Reviews: Drug Discovery 19: 305-306.
- Lassnig C, Sanchez CM, Egerbacher M, Walter I, Majer S, et. al. (2005) Development of a transgenic mouse model susceptible to human coronavirus 229E. Proc Natl Acad Sci USA 102: 8275-8280.
- Wentworth DE, Tresnan DB, Turner BC, Lerman, IR, Bullis B, et. al. (2005) Cells of human aminopeptidase N (CD13) transgenic mice are infected by human coronavirus-229E in vitro, but not in vivo. Virology 335: 185-197. [Crossref]
- Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, et. al. (2014) Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA 111: 4970-4975.
- Agrawal AS, Garron T, Tao X, Peng BH, Wakamiya M, et. al. (2015) Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J Virol 89: 3659-3670.
- Estes JD, Wong SW, Brenchley JM (2018) Nonhuman primate models of human viral infections. Nat Rev Immunol 18: 390-404. [Crossref]
- Zhang Q, Honko A, Zhou J, Gong H, Downs SN, Jhonatan Henao Vasquez JH, et. al. (2020) Cellular Nanosponges Inhibit SARS-CoV 2 Infectivity. Nano Lett 20: 5570-5574.



