The “severe acute respiratory syndrome coronavirus type 2” (SARS-CoV-2) is an enveloped, single-stranded, positive-sense RNA virus, responsible for a highly contagious infection, known as “coronavirus disease 19” (COVID-19). SARS-CoV-2 was discovered in late December 2019. Then, it has spread out from Wuhan, Hubei province, People’s Republic of China, into numerous countries worldwide, and it has caused a global pandemic [1].
A novel variant of SARS-CoV-2, B.1.1.7, originally discovered in the United Kingdom (UK) in August 2020, has rapidly overtaken other strains around the globe [2]. The Applied Bio-Systems TaqPath COVID-19 RT-PCR Kit, ability to detect the S-gene target, was used by the scientists from Portugal. Then, they proceeded the confirmatory test; sequencing, and they found that 91.9 percent of the cases with S- gene failure (SGTF) were the variant of B.1.1.7. Thus, they concluded that S-gene target failure (SGTF) could be a sensitive surrogate of B.1.1.7 [3].
This new variant, SGTF, was estimated to be 40% to 80% more transmissible than previous variants [4] and 35% more lethal than the wild-type virus [5]. And the spread of this variant had led to a surge in COVID-19 cases and deaths in UK. Moreover, in other countries the reproductive number for B 1.1.7 relative to other lineage was higher; 55% in Denmark, 74% in Switzerland, and 59% in the United States [5]
To detect the spread of Spike Gene Target Failure (SGTF) in Yangon, Myanmar, we have been tracking the SGTF prevalence among clinically suspected cases, contact persons and travelers since January 2021 to May 2021. The samples were primarily collected with nasopharyngeal swabs from them. Then, they were screened by using SARS CoV-2 RT-PCR detection kit. After that, all SARS CoV-2 positive samples were tested with “TaqPath-1 Step SARS CoV-2 Detection kit, Thermofisher Scientific”. SGTF was identified as “non-detection of the S gene target among samples that tested positive (cycle threshold less than 37) for both the N gene and ORF1ab gene target” (Figure 1).
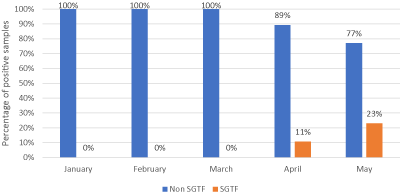
Figure 1. Spike Gene Target Failure according to months in No (1) DSGH
During this period, among 76952 nasopharyngeal swab samples were tested and 623 samples were SARS CoV-2 test positive. The total number of positive samples on January, February, March, April and May were 316, 115, 88, 56, and 48 respectively. In these 623 positive samples, 17 samples (2.7%) were found to have SGTF over five months. Although we have been trying to trace SGTF since January 2021, it was not detected until March 2021. We discovered SGTF only in early April and May 2021 in imported cases only i.e travelers. These imported cases were coming from neighboring ASEAN countries; 8 from India, 2 from Thailand and 7 cases from South Korea. The positivity rate of SGTF over positive SARS CoV-2 was 11% (6/56) in April; it became double in May: 23% (11/48). Thus, the trend of cases with SGTF was increasing and alarming too.
In contrary to the situation in UK; the sudden surge in number of positive SARS CoV-2 cases and increasing mortality with the discovery of new variant- SGTF, Myanmar has different scenario. Although our study was done in military hospitals, the number of confirmed SARS Co V-2 positive cases according to statistics from Ministry of Health and Sports were as follows: 538 cases in March, 383 cases in April and 812 cases in May. Moreover, the number of death cases due to severe COVID-19 infection was as follows; 7 cases in March, 3 cases in April and 8 cases in May. It was clear that the number of both new cases and death cases dropped in April and rose again in May; it was in line with the trend of positivity rate of SGTF. Though we have been doing early detection of cases at entry point to Myanmar and timely quarantine services to imported cases, the increase in rate of new cases and death cases may be due to mutation of SARS CoV-2- SGTF causing rapid transmission.
This study has several limitations. The sequencing technique is not cheap and it is not available in all molecular laboratory; moreover, the reagent for detection of new variant of SARS CoV-2 is expensive in Myanmar. Although these data represent a sample from a region and it highlights the awareness among all health care personnel and publics particularly screening at entry points to Myanmar.
- Guan WJ, Liang WH, Zhao Y, Liang HR, Chen ZS, et al. (2020) Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J 55: 2000547. [Crossref]
- Brown KA, Gubbay J, Hopkins J, Patel S, Buchan SA, et al. (2021) Rapid Rise of S-Gene Target Failure and the UK variant B.1.1.7 among COVID-19 isolates in the Greater Toronto Area, Canada. MedRxiv.
- Borges V, Sousa C, Menezes L, Gonçalves AM, Picão M, et al. (2021) Tracking SARS-CoV-2 lineage B.1.1.7 dissemination: insights from nationwide spike gene target failure (SGTF) and spike gene late detection (SGTL) data, Portugal, week 49 2020 to week 3 2021. ECDC 26: 2100131.
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, et al. (2021) Transmission of SARS-CoV-2 lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. MedRxiv
- Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, et al. (2021) Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science, New York. pp: 372: 6538.

