Abstract
There is a dire need for improved cervical cancer screening methods. New tests are in the pipeline, but their diagnostic capabilities may be limited without a way to assess specimen validity. Here we describe an enzyme-linked immunosorbent assay (ELISA) that captures specific intermediate filament proteins (cytokeratins) from potentially transformable target cells located within or originating from the cervical transformation zone as a means of cervical specimens. Seventy-five uterine cervical samples negative for human papillomavirus (HPV) were grouped based on microscopic analysis for the presence or absence of cervical transformation zone cells, and all samples were tested in both a newly developed recomWell Keratin 5/8/18 ELISA and a pan keratin control ELISA. Additionally, 15 samples from HPV-positive patients manifesting histopathologic lesions or squamous cell carcinoma were tested. Our results demonstrate the presence and detectability by ELISA of keratins 5, 8, and 18 in parabasal, squamous metaplastic, and endocervical cells, while simultaneously suggesting their absence in differentiated squamous cells. We also validate with ELISA the expression of these keratins in HPV-induced disease-state individuals. Our findings indicate recomWell Keratin 5/8/18 ELISA may be useful as a standardizing tool in cervical cancer screening, or alternatively as a quality indicator to denote specimen adequacy.
Key words
cervical cancer screening, cervical transformation zone, keratin, enzyme-linked immunosorbent assay (ELISA), specimen adequacy, standardization
Abbreviations
HPV: Human Papillomavirus; ELISA: Enzyme-Linked Immunosorbent Assay; SCJ: Squamocolumnar Junction; CIN: Cervical Intraepithelial Neoplasia; MEM: Minimum Essential Medium; FCS: Fetal Calf Serum; PBST: Phosphate-Buffered Saline/Tween-20; TMB: Tetramethylbenzidine
Introduction
It has been over thirty years since a link between human papillomavirus (HPV) and cervical cancer was first proposed [1]. This link is now well-established, yet the disease pervades as the fourth most common cancer in women worldwide, with over 500,000 estimated new cases in 2012 [2-3].
Despite the recent introduction of prophylactic HPV vaccines, the quest for improved cervical cancer screening tools continues [4]. Although cervical cytology (Papanicolaou test) has greatly reduced overall morbidity and mortality, and high-risk HPV DNA testing has proven effective when used appropriately, both preventive measures have their own shortcomings [5-8]. In the area of contemporary diagnostics, progress is being made exploiting host cellular biomarkers and viral oncoproteins as potential indicators of HPV-induced cervical lesions and carcinoma. New testing approaches include the use of enzyme-linked immunosorbent assay (ELISA) and immunocytochemistry [9-21]. However, there is relatively sparse literature describing approaches to normalize these tests to ensure sample validity and comparison. Due to the complex nature of both the cervical epithelium and HPV carcinogenesis, and the inherent variance in sample collection methods, it would seem indispensable that a screening test should be accompanied by a sample validity control measure to reduce false negative rates and potential misdiagnosis.
The uterine cervix is subject to an array of physiologic changes over the course of a woman’s lifetime, including those brought about by HPV. The most dynamic region of the cervix is the cervical transformation zone, where ectocervix (stratified squamous epithelium) and endocervix (endocervical columnar and reserve cells) meet, and within which lies the squamocolumnar junction (SCJ). The SCJ is constantly evolving through a process called squamous metaplasia, and the transformation zone defines the changing area between original SCJ and newly formed SCJ [22,23]. HPV is known to infect undifferentiated basal cells, and for reasons not yet fully understood, has a tropism for those found specifically within the cervical transformation zone. In fact, most precancerous lesions and squamous carcinomas originate there. In some cases, persistent infection leading to overexpression of viral oncoproteins E6 and E7 may inhibit these cells from normal mitotic arrest and entry into squamous cell differentiation, and instead drive them to proliferate abnormally through stages of cervical intraepithelial neoplasia (CIN) and ultimately cancer [24].
Based on this knowledge, one strategy for would be to find a molecular marker detectable only in undifferentiated cells within or originating from the cervical transformation zone. Any detection of endocervical cells could also verify whether the transformation zone has been properly sampled, echoing specimen quality guidelines outlined by Pap cytology [22,25,26]. Critical would be the absence of the marker from commonplace cervical components such as immune cells, erythrocytes, and normal bacterial flora. Human cytokeratins, found only in epithelial cells, are intermediate filament proteins manifesting high molecular diversity [27]. Although their biology is complex and over 50 functional keratin genes exist, studies have shown general expression patterns in normal cervical epithelium [27]. ”Maturation” keratins normally present in differentiated intermediate and superficial squamous cells include keratins 4, 10, and 13, while stably expressed keratins localized to undifferentiated basal/parabasal/reserve, squamous metaplastic, and endocervical columnar cells include keratins 5, 8, and 18 [28-32].
A validation marker’s expression should be unaffected by pathology. In clinical applications, keratins have evolved to become one of the most potent markers of differentiation in the diagnosis of epithelial tumors because epithelial tumors and metastases usually retain the keratin patterns of their epithelial origin [27]. However, few studies have looked specifically at possible keratin changes in precursor CIN or cervical squamous cell carcinoma. Despite this literary deficiency, some studies suggest that there is no phenotypic loss of keratins 8, 18, and/or 5 in both tumors and CIN lesions [28-30,32-36].
The intent of this study was to determine if ELISA detection of keratins 5, 8, and 18 in cervical specimens is not only but also practical as a method for validation of cervical samples scheduled for cervical cancer screening assays. Seventy-five HPV-negative uterine cervical samples were grouped based on microscopic analysis for the presence or absence of cervical transformation zone cells, and all samples were tested in both a newly developed recomWell Keratin 5/8/18 ELISA and a pan keratin control ELISA. An additional 15 samples from HPV-positive patients with confirmed histopathology or squamous cell carcinoma were tested. Our results suggest that the recomWell Keratin 5/8/18 ELISA may serve a useful role as a standardizing tool in future diagnostics.
Materials and methods
Antibodies
Pan keratin sandwich ELISA: Mouse monoclonal anti-pan keratin antibody (Cytokeratins 4, 5, 6, 8, 10, 13, 18) (Cell Signaling Technology, Danvers, Massachusetts USA) was used for capture. Biotinylated mouse monoclonal anti-pan keratin antibody (4, 5, 6, 8, 10, 13, 18) (Cell Signaling Technology) was used for detection.
ELISA: Keratin 5/8/18 sandwich ELISA: For detection of keratin 5, 8, and 18, the recomWell Keratin 5/8/18 ELISA (Mikrogen GmbH, Neuried, Germany) was used.
Cell lines
All cell lines were derived from cervical carcinoma. MS751 (HTB-34) and ME-180 (HTB-33) cell lines were purchased from America Type Culture Collection. MS751 cells were maintained in ATCC formulated Eagle’s Minimum Essential Medium (MEM), with 10% fetal calf serum (FCS) and 1 mM sodium pyruvate. ME-180 cells were maintained in ATCC formulated McCoy’s 5A Modified Medium, with 10% FCS; HeLa (CCL2) cell line was purchased from DSMZ (Braunschweig, Germany), and maintained in Eagle’s MEM, supplemented with 10% FCS. Cerv-215 cell line was purchased from Cell Lines Service (Eppelheim, Germany) and maintained in Eagle’s MEM, supplemented with 10% FCS, 1 mM sodium pyruvate, and 1% non-essential amino acids. All cell lines were additionally supplemented with antibiotics (1% penicillin/streptomycin) and cultured in 5% CO2 at 37°C. Mycoplasma testing was performed using the Venor GeM One Step Kit from Minerva Biolabs.
Clinical sample collection and processing
The study was approved by ethics committees from participating institutions, informed consent was obtained from all individuals, and testing was performed in accordance with the principles of the Declaration of Helsinki. Samples were obtained from the Charite Hospital (Berlin, Germany) and the IV. Clinic of Obstetrics and Gynaecology, Hippokration Hospital – Aristotle University of Thessaloniki (AUTH) (Thessaloniki, Greece). Additional samples were received from Klinikum Wolfsburg (Wolfsburg, Germany). Data from diagnostic testing done in both Greece and Germany was entered into a Data Capture System database for easy retrieval by all parties involved. Data is catalogued in Appendix A (Supplemental Digital Content).
Collection
Cervical epithelium samples were collected from patients using first a Cervex-Brush, and then a cytobrush. Both brushes were put into a ThinPrep vial containing 20 ml of PreservCyt Solution. Adequate amounts were then extracted for performing liquid-based cytology (Cytology Lab, AUTH) and HPV testing (Gynecologic Tumor Immunology Lab, Charite-Universitaetsmedizin Berlin). The remaining volume was extracted to a 15 ml Falcon tube and sent to Mikrogen (Neuried, Germany) (ambient temperature), where samples were stored at 4°C until testing in keratin ELISAs. *A subset of 6 samples (Group F) from the Charite was obtained under an alternative method: Two consecutive cytobrush samples were taken during colposcopic examination. The first was used for cytology and HPV testing, as described below. The second was placed directly into a sample buffer (0.1% Tween-20, 1x phosphate-buffered saline (PBST)) and frozen at -80°C. To prepare lysates, samples were thawed and 20 µl of 50x protease inhibitor stock solution (Roche Diagnostics, Germany) was added per tube. Excess liquid was wiped from the brush, and the brush was discarded. Sample was centrifuged (20 min, 13,000 rpm, 4°C) and the supernatant was aliquoted to a new tube and kept frozen at -20°C until keratin ELISA testing. **Klinikum Wolfsburg samples were collected in ThinPrep with a Cervex-Brush, and both cytology and Hybrid Capture 2 (Digene, Gaithersburg, Maryland) HPV testing were performed on-site before being sent to Mikrogen.
Cytology
Slides were prepared using a ThinPrep without space 2000 Processor (Hologic), followed by fixation and standard Papanikolaou staining, and final examination by a certified cytopathologist. HPV Testing: HPV genotyping was done by HPV broad spectrum multiplex genotyping [37]. This method amplifies a sequence from the L1 gene by generic primers GP5+/GP6+ and uses type-specific probes coupled to Luminex beads, allowing for the detection of multiple high- and low-risk HPV types in a single reaction. For samples collected at AUTH, 1 ml of ThinPrep sample was transferred to an Eppendorf tube and centrifuged (6000 rpm, 5 min). The supernatant was discarded, and the pellet was stored at -20°C until shipment to Charite Tumor Immunology Lab by ordinary mail at ambient temperature.
Colposcopy/Histology
Women with either cytology result positive (atypical squamous cells of undetermined significance or worse) or HPV DNA test positive (high-risk types only) were called back for colposcopy. If warranted, biopsy was taken and histological analysis was done at the Charite Institute for Pathology and the Department of Histology, Hippokration Hospital of Thessaloniki, Greece. Pathology Coding: Scoring for cytology, colposcopy, and histology was done based on the ASSIST approach, as described by Agorastos, et al. [38]. This modification method attempts to unify multiple patient record repositories in geographically different locations, allowing for common interpretation and the formation of “on demand” study groups.
Hematoxylin and Eosin staining and microscopy
Samples were pre-selected for staining based on the results of prior diagnostic testing. Each sample (15 ml Falcon tube) was vortexed at moderate speed and two separate 10 µl drops were added to a standard microscope slide (Carl Roth, Karlsruhe, Germany). The slide was air dried under a fume hood, then heat-fixed by running it through a Bunson burner flame three times in quick succession. Once cool, slides were flooded with Mayer’s Hematoxylin (Suesse, Gudensberg, Germany) for 30 seconds, and then dipped into a staining chamber containing tap water for 30 seconds. Slides were then covered with 95% ethanol for 5 seconds, followed by a quick rinse with fresh tap water. Finally, aqueous Eosin (Suesse) was added to slides for 15 seconds, and slides were rinsed in another tap water chamber for 10 seconds. Slides were ultimately visualized with a light microscope (Zeiss, Munich, Germany). A Canon digital camera was used to generate images.
Grouping of samples
HPV-negative specimens were grouped accordingly based on microscopy. Samples showing at least the presence of squamous basal/parabasal, squamous metaplastic, and/or endocervical columnar cells fell into Group A. Samples showing only intermediate and/or superficial squamous cells on smear were placed into Group B. Ambiguous samples that did not meet the aforementioned criteria were excluded from this study. HPV-positive, histopathology-positive specimens consisted of additional Groups C (ThinPrep) and F (freeze-lysed in buffer). All sample volumes other than those in Group F ranged from 6.5 to 20 ml. Visual characteristics of sample pellets were documented when possible. Using colored water, a range of volumes was pipetted into 15 ml Falcon tubes to serve as a reference for estimating sample pellet size (in microliters). Patient age range was 20-71.
Lysis of cell lines and clinical samples
Cell lines and clinical specimens were centrifuged at 600 x g for 15 minutes. ThinPrep supernatant was discarded and 500 µl Lysis Buffer A (Mikrogen GmbH, Neuried, Germany) was added to scratched pellet. Tubes were vortexed and allowed to sit at room temperature for 30 minutes before the addition of 500 µl of Lysis Buffer B (Mikrogen GmbH, Neuried, Germany). Cell lines were further diluted in a 1:1 solution of Lysis Buffer A and Lysis Buffer B to give preferred concentrations for testing. All samples were tested in the pan keratin control ELISA and the keratin 5/8/18 ELISA, as described below. The 1:1 Lysis Buffer solution also served as negative control in all assays.
Pan keratin control sandwich ELISA*
Pan keratin (capture) antibody diluted in coating buffer (Mikrogen GmbH, Neuried, Germany) to 0.05 µg/ml was added (100 µl/well) to a 96-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany), and the plate was incubated overnight at 4°C. Wells were washed 3 times with Washing Buffer (Mikrogen GmbH, Neuried, Germany). Then 300 µl Blocking Buffer (Mikrogen GmbH, Neuried, Germany) was added to each well, and the plate was incubated for 2 hours at room temperature. Wells were aspirated and allowed to dry for 2 hours at 30°C. Addition of 100 µl of lysed sample to each well was followed by a 1-hour incubation at room temperature. The plate was washed 3 times with Washing Buffer (Mikrogen GmbH, Neuried, Germany). Biotinylated pan keratin (detection) antibody was diluted in Conjugate Buffer (Mikrogen GmbH, Neuried, Germany) to 0.05 µg/ml, and 100 µl was added to each well. This was followed by another 1-hour incubation at room temperature. Wells were washed 3 times again with Washing Buffer. Then 100 µl of Streptavidin-conjugate (recomWell Keratin 5/8/18, Mikrogen GmbH, Neuried, Germany) was added to each well. The plate was incubated for 1 hour at room temperature, succeeded by 6 washes with washing buffer. One hundred microliters of tetramethlybenzidine (TMB) detection reagent was added to each well, and the plate was incubated for 30 minutes in the dark. Finally, 100 µl stop solution (24.9% H3PO4) was added to each well, and absorbance was measured at 450nm on a Berthold Mithras LB940 plate reader (Berthold Technologies, Bad Wildbad, Germany).
Keratin 5/8/18 sandwich ELISA*
The samples were tested in multiple recomWell Keratin 5/8/18 (Mikrogen GmbH, Neuried, Germany) assays. In particular: 100 µl of lysed sample was added to each well, followed by a 1-hour incubation at room temperature. The plate was washed 3 times with Washing Buffer. Then 100 µl of the detection antibody was added to each well. This was followed by another 1-hour incubation at room temperature. Wells were washed 3 times again with Washing Buffer, and 100 µl of the Strepatividin conjugate was then added to each well. The plate was incubated 1 hour at room temperature, succeeded by 6 washes with Washing Buffer. Then 100 µl of TMB detection reagent was added to each well, and the plate was incubated for 30 minutes in the dark. Finally 100 µl Stop Solution was added to each well, and absorbance was measured at 450nm on a plate reader (Berthold Mithras LB940).
*Both pan keratin and recomWell Keratin 5/8/18 assays described herein pertain to cell line testing. For details concerning the testing of clinical samples, see Appendix A.
Results
Our initial focus was to determine if keratins 5, 8, and 18 could be detected in an ELISA. To test this, a sandwich ELISA – the recomWell Keratin 5/8/18 - employing monoclonal mouse antibodies specific for keratins 5, 8, and 18 was created. Additionally, a pan keratin assay capable of detecting human cytokeratins 4, 5, 6, 8, 10, 13, and 18 was developed for use as a control in subsequent clinical sample testing. Four cervical carcinoma cell lines were titrated and run in both assays. Figure 1 shows clear detection of proteins in three cell lines (Cerv-215, MS-751, HeLa) in the recomWell Keratin 5/8/18 (Figure 1A) and the pan keratin ELISA (Figure 1B) to 1000 cells per well. These results are supported by known keratin expression profiles (provided by the manufacturers) for at least HeLa and Cerv-215 cell lines, which include keratins 8 and 18. Moreover, no background “noise” was observed in either assay, as absorbance (OD450) values of 0 were obtained for buffer controls in both tests.
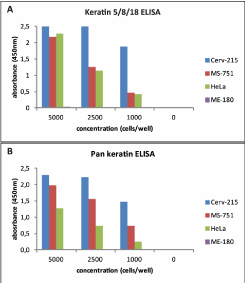
Figure 1. Cell line testing Keratin detection in cervical carcinoma cell lines using two different enzyme-linked immunosorbent assays (ELISAs).
As cell lines have lim2021 Copyright OAT. All rights reservterine cervical samples for in vivo keratin expression, with the hope of answering the question of whether or not keratins 5, 8, and 18 can be detected collectively by ELISA in true clinical specimens. Hematoxylin and Eosin staining was done on HPV-negative samples, and they were divided into two groups based on microscopic observations. Those in the first (Group A) exhibited cell types in which keratins 5, 8, and/or 18 were likely to be found (Figure 2). These include parabasal (Figure 2A), squamous metaplastic (Figure 2B), and endocervical columnar (Figure 2C) cells. The images are represented by actual samples (A23, A2, A8, respectively) tested in both the recomWell Keratin 5/8/18 and pan keratin assays (Figure 2D). Prior testing with HeLa cells (unpublished data) allowed for optimization of antibody coating concentrations for each assay. However, detection concentrations used in the various recomWell 5/8/18 assay systems varied slightly from sample to sample (see Appendix A). Nonetheless, using a generous arbitrary threshold value (as all experimental blanks were 0) of OD450=0.1, it was shown that 28 of the 32 samples (87.5%) manifesting any of the above described cell types on smear gave positive signals (OD450 ≥0.1, mean = 0.98) in the recomWell Keratin 5/8/18, thus showing that these keratins can be detected in clinical samples with a sandwich ELISA. These 28 samples also showed positive results in the pan keratin assay. Of the 4 negative recomWell Keratin 5/8/18 samples, 3 of them (A17, A20, A30) showed strong pan keratin signals (minimum OD450 of 1.47) compared to the fourth (A11), which showed a weak positive pan keratin signal of OD450=0.20. Noteworthy is that not all Group A samples contained exclusively target cells, as a number of them also showed differentiated squamous cells, like those shown in Figure 2A.
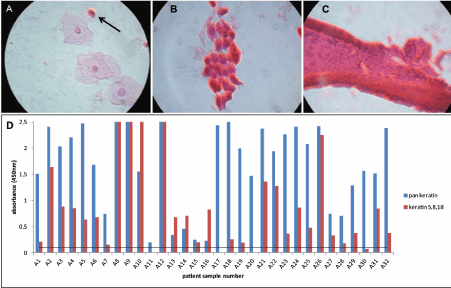
Figure 2. Microscopy and ELISA results for Group A (A) Sample A23 shows a single parabasal cell in the presence of superficial squamous cells (1000x magnification). (B) Squamous metaplastic cells as seen in sample A2 (1000x). (C) A cluster of endocervical columnar cells as seen in sample A8 (1000x). (D) Widespread detection in the recomWell Keratin 5/8/18 ELISA, suggesting these specific keratins are measurable by ELISA in clinical specimens.
To demonstrate the suspected absence of keratins 5, 8, and 18 in differentiated cells of the stratified squamous epithelium, HPV-negative samples showing only intermediate and/or superficial cells on microscopy were categorized as Group B and tested in both assays (Figure 3). Of extra importance is that some of these specimens also contained potential interference factors commonly found in the cervical environment, such as immune cells (Figure 3A), bacteria (Figure 3B), and blood (Figure 3C). Using the same threshold value of OD450=0.1 assigned for Group A, 27 out of 43 Group B samples (62.8%) were negative in the recomWell Keratin 5/8/18, including those represented in Figures 3A, 3B, and 3C (samples B37, B7, B8, respectively). Meanwhile, 25 of these 27 recomWell Keratin 5/8/18 negatives were positive in the pan keratin assay. This signifies, in concordance with microscopy, that for a majority there are cervical cells (intermediate, superficial) present in the sample (verified by a positive pan keratin assay capable of detecting “maturation” keratins), but they are not producing keratins 5, 8, and/or 18 (negative recomWell Keratin 5/8/18 result). Surprisingly, one sample (B19) demonstrated a negative pan keratin result and a positive recomWell Keratin 5/8/18 result.
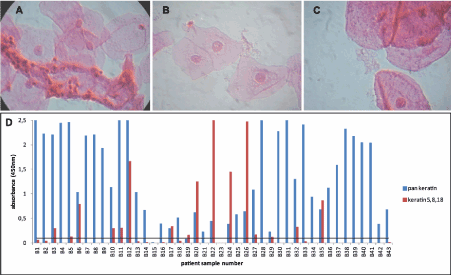
Figure 3. Microscopy and ELISA results for Group B (A) An infiltrate of leukocytes masking superficial epithelial cells as seen in sample B37 (1000x). (B) Intermediate cells surrounded by numerous bacilli in sample B7 (1000x). (C) Sample B8 shows red blood cell ghosts mixed in with superficial cells (1000x). Visual inspection showed a bloody sample pellet. (D) 25 of the 27 samples negative in the recomWell Keratin 5/8/18 ELISA are positive in the pan keratin ELISA, thus demonstrating the likely absence of keratins 5, 8, and 18 in differentiated squamous cells.
Our final interest was in detecting keratins 5, 8, and 18 in samples derived from HPV-positive patients with histopathology. Therefore, these samples (Groups C and F) were run in both assays (Figure 4). Group C samples were collected in ThinPrep, while Group F samples were collected in PBST and frozen before use. Microscopy was done on Group C samples, but was not possible on Group F. All 9 Group C samples showed at least high-grade ASSIST histology scores (>=2) except one (C4), which lacked histology data (see Appendix A for grading scale). However, colposcopy for sample C4 showed a value of 3 (indicative of invasive carcinoma) so it was included. In the recomWell Keratin 5/8/18 (Figure 4A), one sample (C5) showed a strong signal (OD450>2), while 5 (C1-C4, C6) samples showed weak to moderate signals (OD450 0.1-0.5). Three samples (C7-C9) showed no signal in the recomWell Keratin 5/8/18 test. All 6 Group F samples (1 low-grade, 5 high-grade) displayed moderate to strong signals in the recomWell Keratin 5/8/18 assay (Figure 4B). These results show keratins 5, 8, and 18 are detectable by ELISA in patients manifesting HPV-induced histopathologic lesions and invasive cancer. All samples from both groups showed moderate to strong signals in the corresponding pan keratin assay.
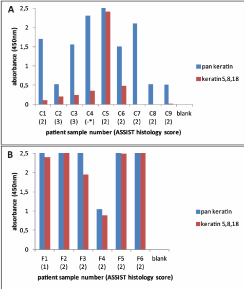
Figure 4. Keratin detection in human papillomavirus (HPV)-positive patients with confirmed histopathology ASSIST scoring: 1= low-grade, 2= high-grade, 3= suspicious for invasive carcinoma (A) ThinPrep-preserved Group C samples show a range of signals in the recomWell Keratin 5/8/18 ELISA. *Sample C4 lacked histology data but showed a colposcopy score of 3, suggesting cancer. (B) Group F samples, preserved in buffer (0.1% Tween-20, 1x phosphate-buffered saline) and freeze-lysed before use, show strong signals in both assays. Conclusively, keratins 5, 8, and 18 are detectable by ELISA in clinical samples collected and prepared under two different methods.
Discussion
There is a growing need for improved cervical cancer screening methods worldwide. Cytology has been the backbone of diagnostics for 50 years, but its poor sensitivity rate makes it a far from perfect test [7]. Adequate sample collection is a prerequisite for any screening test to be consistently informative. Promising new test approaches are in development, but their diagnostic capabilities may be limited without a way to assess specimen validity.
In this paper, we address this latter issue by proposing to capture keratins from potentially transformable target cells located within or originating from the cervical transformation zone in an ELISA as a means of validating clinical specimens. To the best of our knowledge, no immunoassays have ever been developed attempting to survey in combination the specific keratins 5, 8, and 18 normally expressed by these cells. Our initial testing with cervical cancer cell lines showed that measurement is technically possible. However, expression patterns vary from cell line to cell line. Furthermore, cultured cell lines cannot mimic real cells of the cervical epithelium, where intrinsic factors such as hormones play a role in dictating events like differentiation, and above all, expression levels of proteins such as keratins [23]. It is generally unknown exactly how much keratin 5, 8, and 18 is expressed in vivo under normal physiologic conditions, and if these levels are even quantifiable in an assay. Therefore, the true test for keratin detection would be with actual clinical samples.
Microscopy allowed for pre-characterization of HPV-negative clinical specimens based on cell content before running any assays. By doing this, we could then anticipate, based on literary knowledge of cervical keratin expression patterns, the type of result each sample might produce in both the recomWell Keratin 5/8/18 and pan keratin assays. If a keratin 5/8/18 signal were to be obtainable by ELISA, then samples that showed the presence of parabasal, squamous metaplastic, and/or endocervical cells on smear (Group A) would be the best candidates to demonstrate this phenomenon. Indeed, 28 of the 32 (87.5%) Group A samples did show positive results in the recomWell Keratin 5/8/18. Of utmost importance is that a number of these positive samples also showed superficial cells on microscopy, thereby suggesting no sample bias from patients displaying conditions such as squamous atrophy. In regard to the recomWell Keratin 5/8/18 negatives, it is likely that even though required cells were seen on smear, there weren’t enough of them in the sample to produce a measurable result. Three of these (A17, A20, A30) gave strong positive pan keratin results, but as many superficial/intermediate cells were seen on smear, this result seems logical. In contrast, the fourth negative (A11) showed very few cells of any kind on smear, and subsequently yielded a weak pan keratin result too. In summary, the ELISA results correlated well with the visual impression gained by microscopic observation of the samples.
To show any recomWell Keratin 5/8/18 signals were not being produced from unwanted differentiated squamous cells, samples that presented only superficial and/or intermediate cells on microscopy (Group B) were also tested. Strategically, some samples were used that also included common cervical components (leukocytes, bacteria, blood) that could potentially cause assay interference, leading to background noise or even false positives. The pan keratin assay, capable of detecting not only basal but also maturation keratins (such as 4, 10, and 13), served as a verification control (to microscopy) to confirm the overall presence of cells in the sample in cases where recomWell Keratin 5/8/18 was negative. Of the 43 Group B samples tested, 27 were recomWell Keratin 5/8/18 negative (62.8%). On the other hand, 25 of those 27 were pan keratin positive. This result pattern favors the notion that differentiated cells do not produce keratins 5, 8, and 18. Some samples (such as B37, B7, and B8) exhibiting potential interference factors on the smear showed zero signal in the recomWell Keratin 5/8/18. Thus, no background interference occurs as a result of these components being present. One may offer the opposite theory in that these components may actually be inhibiting the production of signal from other cells in the sample. However, the evidence refutes this possibility. For example, sample A9 was bloody and also showed many immune cells (in addition to basal cells), yet still produced a very strong recomWell Keratin 5/8/18 signal. Also, pan keratin signals for samples B37, B7, and B8 were all positive even though recomWell Keratin 5/8/18 results were negative. Ultimately, however, the question arises as to why 16 samples were 5/8/18 positive when no “indicator” cells were seen on smear. At best, microscopy is not perfect, and only a small amount of total sample was used for analysis. It is probable that these cells were either missed on microscopic observation, or were absent from the aliquots used to make the slide altogether but present in the main sample itself. We cannot totally discount the hypothesis that differentiated cells also contain questionable amounts of keratins 5, 8, and/or 18, but based on the evidence generated from this and other studies, it is unlikely. Two samples (B15, B23) produced negative results in both keratin assays while showing many superficial cells on smear. These results cannot be explained and would count as false negatives. Finally, sample B19 showed a negative pan keratin ELISA result but a positive recomWell Keratin 5/8/18 result. However, as both OD450 values (pan = 0.08, recomWell Keratin 5/8/18 = 0.15) hover near threshold, we consider these discordant results inconclusive.
Interesting to note is that specimen pellet size seemed to play no role in predicting specimen adequacy. For example, A7 had a pellet of 10 µl and a recomWell Keratin 5/8/18 OD450 of 0.15. Sample A4 had a pellet of less than 10 µl, yet produced an OD450 of 0.85. The former hovers around threshold value, but the latter shows a moderately strong signal, even though both pellet sizes are roughly the same. Samples B4 and B7, with respective pellet sizes of 160 µl and 180 µl, both gave pan keratin signals of OD450>2.0. However, both also gave an OD450 value of zero in the recomWell Keratin 5/8/18. Our proposed sample validation method would suggest B4 and B7 are thus inadequate samples, even though one ignorant of the assay results may be tempted to say otherwise simply because there is much visible material in the tube. We show here that quality is superior to quantity. This ideology runs contrast to a normalization method used by OncoHealth Corporation (Fremont, California, USA) in their commercially available Whole Cell ELISA for the detection of HPV oncoproteins E6 and E7. Their concept is based on cellularity, insinuating all samples can be standardized using equivalent pellet to volume ratios. The pitfall here is that it dangerously assumes all content in a pellet is cervical cells. Even if this were true, these cells may not be the select (undifferentiated) target cervical cells required for detection of high levels of E6 and E7 proteins. In effect, it fails to acknowledge the fact that there will almost always be other non-cervical cells and mucus in a sample, sometimes in excess quantity. Without a way to distinguish the true nature of a sample, relying on a cellularity method is tantamount to flipping a coin.
In this study, we also tested samples collected from HPV-positive patients manifesting histopathology. Although we could show keratins 5, 8, and/or 18 are measurable in some patients, definitive results are not possible from this study alone. If we assume proper sample collection in every case, it is confounding why samples with advanced lesions (C1, C4, C6-C9) or even cancer (C2, C3) would show such low levels of keratins 5, 8, and 18. One would be inclined to expect the opposite, as dysplastic dyplastic cells harboring these basal keratins should increase with disease progression [24,29,30,32]. Nevertheless, we do see cases where proper samples have not necessarily been obtained. Samples C7-C9 showed only superficial cells and no clear dysplastic or transformation zone cells on microscopy. Microscopy herein refers to that done at Mikrogen and in some cases conflicts with results obtained from cytology performed at the clinical laboratories. Indeed, there are even cases of HPV-negative samples with cytology ASSIST scores of 1 (mild findings) or 2 (severe findings), but subsequent colposcopy and/or histology testing shows only low-grade or insignificant findings (see Appendix A). Due to the relative inter-observer subjectivity and variability associated with cytology, HPV-induced “pathology” in this study was therefore defined by the “gold standard” of histological diagnosis. Pan keratin results were positive, but recomWell Keratin 5/8/18 results were negative, inferring inadequacy. Considering this, it is difficult to gauge any effect pathology might have on keratin expression without running a parallel diagnostic test for elevated oncoprotein or host biomarker of interest on the same sample. This is something that needs further exploration. What we can say is that, without concomitant assessment of sample validity, samples such as C7, C8, and C9 might generate false negative results if tested alone for said oncoprotein or biomarker. Additionally, there seemed to be a discrepancy in results between Groups C and F. Whereas Group C ThinPrep samples showed generally low 5/8/18 signals, Group F samples (collected in PBST and freeze-lysed), from patients with both low- and high-grade lesions, all showed strong signals. This difference is somewhat puzzling. Microscopy could not be done on Group F samples, so there was no way to deduce cell content. Regardless, results show keratins 5, 8, and 18 can be detected using multiple sample collection/lysis methods.
The recomWell Keratin 5/8/18 ELISA has potential to serve as a test for sample validation in a range of cervical cancer screening tests. Future prospects include ELISAs developed for detection of elevated tumor suppressor protein p16, and HPV oncoprotein E7 [11,21,39,40]. A dual staining cytology technique for p16 and Ki-67 shows much promise [12]. Further diagnostic tests exploiting overexpressed proteins such as ProExC, COX-2, CXCL12, p63, and survivin are in the pipeline [13,14,17,19]. However, other than the OncoHealth assay already described, the only new attempts to address the issue of validation seem to come alongside the E7 assay development. Ehehalt and colleagues use a DNA-binding dye (Eva Green) approach to estimate cell content in a sample [21]. A major flaw in this strategy is that the dye will bind not only keratinocyte DNA but also the DNA of immune cells and bacteria found normally within the cervix. Prior experiments at Mikrogen (data unpublished) showing strong Eva Green fluorescence detection in E. coli have proven this. Polymerase chain reaction-based techniques for HPV testing have continuously struggled to find a suitable companion reference gene for normalization [39]. Regardless, HPV testing comes with an inherently different strategy in that HPV DNA, as viral load permits, can generally be found in all layers of the stratified cervical epithelium in infected patients.
Conclusion
In conclusion, we have demonstrated the ability to capture keratins 5, 8, and 18 in a sandwich ELISA using healthy and disease-state cervical specimens. Nevertheless, more work needs to be done. A major shortcoming of this study is the low number of pathology samples used. Many more, preferably in parallel with a diagnostic screening test such as that for HPV E7, must be tested to elucidate any kind of effect on keratin expression or correlation between the two subjects measured. Unlike our study, which used variable concentrations of test components, future samples should all be run on the same assay platform as well. Although it was partially addressed herein with microscopy and visual pellet inspection, more direct specificity studies are recommended. Finally, as multiple keratin proteins can be detected from multiple cell types and signal origin remains ambiguous, making a leap from a semi-quantitative to a quantitative assay may prove challenging. Despite this, we believe the recomWell Keratin 5/8/18 ELISA has great potential to serve as a test for sample validation in cervical cancer screening, or, at the very least, as a reliable quality indicator to denote specimen adequacy.
Funding
Funding for this project was received from the following source: EU-Project PIPAVIR (FP7, Grant Agreement Number: 304927).
Conflict of interest
A patent application concerning the subject matter described herein has been filed with the European Patent Office.
References
- Dürst M, Gissmann L, Ikenberg H, zur Hausen H (1983) A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci U S A 80: 3812-3815. [Crossref]
- zur Hausen H (2009) Papillomaviruses in the causation of human cancers - a brief historical account. Virology 384: 260-265. [Crossref]
- International Agency for Research on Cancer, World Health Organization (2012) Cervical Cancer: Estimated Incidence, Mortality and Prevalence Worldwide in 2012.
- Tyler M, Tumban E, Chackerian B (2014) Second-generation prophylactic HPV vaccines: successes and challenges. Expert Rev Vaccines 13: 247-255. [Crossref]
- Schiffman M, Wentzensen N, Wacholder S, Kinney W, Gage JC (2011) Human Papillomavirus Testing in the Prevention of Cervical Cancer. J Natl Cancer Inst 103: 368-383. [Crossref]
- Wright TC Jr, Schiffman M, Solomon D, Cox JT, Garcia F, et al. (2004) Interim guidance for the use of human papillomavirus DNA testing as an adjunct to cervical cytology for screening. Obstet Gynecol 103: 304-309. [Crossref]
- Boone JD, Erickson BK, Huh WK (2012) New insights into cervical cancer screening. J Gynecol Oncol 23: 282-287. [Crossref]
- Rebolj M, Pribac I, Frederiksen ME, Lynge E (2013) The problem of false-positive human papillomavirus DNA tests in cervical screening. Curr Pharm Des 19: 1439-1449. [Crossref]
- Klaes R, Friedrich T, Spitkovsky D, Ridder R, Rudy W, et al. (2001) Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer 92: 276-284. [Crossref]
- Mimica M, Tomi� S, Kardum G, Hofman ID, Kaliterna V, et al. (2010) Ki-67 quantitative evaluation as a marker of cervical intraepithelial neoplasia and human papillomavirus infection. Int J Gynecol Cancer 20: 116-119. [Crossref]
- Balasubramanian A, Hughes J, Mao C, Ridder R, Herkert M, et al. (2009) Evaluation of an ELISA for p16INK4a as a screening test for cervical cancer. Cancer Epidemiol Biomarkers Prev 18: 3008-3017. [Crossref]
- Ikenberg H, Bergeron C, Schmidt D, Griesser H, Alameda F, et al. (2013) Screening for cervical cancer precursors with p16/Ki-67 dual-stained cytology: results of the PALMS study. J Natl Cancer Inst 105: 1550-1557. [Crossref]
- Beccati MD, Buriani C, Pedriali M, Rossi S, Nenci I (2008) Quantitative detection of molecular markers ProEx C (minichromosome maintenance protein 2 and topoisomerase IIa) and MIB-1 in liquid-based cervical squamous cell cytology. Cancer 114: 196-203. [Crossref]
- Zhou WQ, Sheng QY, Sheng YH, Hou WJ, Xu GX, et al. (2015) Expressions of survivin, P16(INK4a), COX-2, and Ki-67 in cervical cancer progression reveal the potential clinical application. Eur J Gynaecol Oncol 36: 62-68. [Crossref]
- Yang YS, Smith-McCune K, Darragh TM, Lai Y, Lin JH, et al. (2012) Direct Human Papillomavirus E6 Whole-Cell Enzyme-Linked Immunosorbent Assay for Objective Measurement of E6 Oncoproteins in Cytology Samples. Clin Vaccine Immunol 19: 1474-1479. [Crossref]
- Sarmadi S, Izadi-mood N, Pourlashkari M, Yarandi F, Sanii S (2012) HPV L1 capsid protein expression in squamous intraepithelial lesions of cervix uteri and its relevance to disease outcome. Arch Gynecol Obstet 285: 779-784. [Crossref]
- Cheung AN, Tsun KL, Ng KM, Szeto E, Siu MK, et al. (2010) P634A4 and TAp73 immunocytochemistry in liquid-based cervical cytology--potential biomarkers for diagnosis and progress prediction of cervical neoplasia. Mod Pathol 23: 559-566. [Crossref]
- Goto T, Takano M, Sasa H, Tsuda H, Yamauchi K, et al. (2006) Clinical significance of immunocytochemistry for PIK3CA as a carcinogenesis-related marker on liquid-based cytology in cervical intraepithelial neoplasia. Oncol Rep 15: 387-391. [Crossref]
- Jaafar F, Righi E, Lindstrom V, Linton C, Nohadani M, et al. (2009) Correlation of CXCL12 expression and FoxP3+ cell infiltration with human papillomavirus infection and clinicopathological progression of cervical cancer. Am J Pathol 175: 1525-1535. [Crossref]
- Ghittoni R, Accardi R, Hasan U, Gheit T, Sylla B, et al. (2010) The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes 40: 1-13. [Crossref]
- Ehehalt D, Lener B, Pircher H, Dreier K, Pfister H, et al. (2012) Detection of Human Papillomavirus Type 18 E7 Oncoprotein in Cervical Smears: a Feasibility Study. J Clin Microbiol 50: 246-257.
- Cibas ES, Ducatman BS (2014) Cytology: Diagnostic Principles and Clinical Correlates, fourth ed., Saunders, Philadelphia PA.
- Jordan J, Singer A (2006) The Cervix, second ed., Wiley-Blackwell, Malden MA.
- Doorbar J, Egawa N, Griffin H, Kranjec C, Murakami I (2015) Human papillomavirus molecular biology and disease association. Rev Med Virol 25: 2-23. [Crossref]
- Solomon D, Nayar R (2004) The Bethesda System for Reporting Cervical Cytology, second ed., Springer-Verlag Publishers, New York NY.
- Zhao C, Austin RM (2008) Adjunctive high-risk human papillomavirus DNA testing is a useful option for disease risk assessment in patients with negative Papanicolaou tests without an endocervical/transformation zone sample. Cancer 114: 242-248. [Crossref]
- Moll R, Divo M, Langbein L (2008) The human keratins: biology and pathology. Histochem Cell Biol 129: 705-733. [Crossref]
- Ivanyi D, Groeneveld E, Van Doornewaard G, Mooi WJ, Hageman PC (1990) Keratin subtypes in carcinomas of the uterine cervix: implications for histogenesis and differential diagnosis. Cancer Res 50: 5143-5152. [Crossref]
- Smedts F, Ramaekers F, Troyanovsky S, Pruszczynski M, Link M, et al. (1992) Keratin expression in cervical cancer. Am J Pathol 141: 497-511. [Crossref]
- Smedts F, Ramaekers F, Troyanovsky S, Pruszczynski M, Robben H, et al. (1992) Basal-cell keratins in cervical reserve cells and a comparison to their expression in cervical intraepithelial neoplasia. Am J Pathol 140: 601-612. [Crossref]
- Purkis PE, Steel JB, Mackenzie IC, Nathrath WB, Leigh IM, et al. (1990) Antibody markers of basal cells in complex epithelia. J Cell Sci 97: 39-50. [Crossref]
- Nair SA, Nair MB, Jayaprakash PG, Rajalekshmy TN, Nair MK, et al. (1997) Increased expression of cytokeratins 14, 18 and 19 correlates with tumor progression in the uterine cervix. Pathobiology 65: 100-107. [Crossref]
- C Carrilho, L Cirnes, M Alberto, L Buane, N Mendes, et al. (2005) Distribution of HPV infection and tumour markers in cervical intraepithelial neoplasia from cone biopsies of Mozambican women. J Clin Pathol 58: 61-68. [Crossref]
- Carrilho C, Alberto M, Buane L, David L (2004) Keratins 8, 10, 13, and 17 are useful markers in the diagnosis of human cervix carcinomas. Hum Pathol 35: 546-551. [Crossref]
- Puts JJ, Moesker O, Kenemans P, Vooijs GP, Ramaekers FC (1985) Expression of cytokeratins in early neoplastic epithelial lesions of the uterine cervix. Int J Gynecol Pathol 4: 300-313. [Crossref]
- Martens J, Baars J, Smedts F, Holterheus M, Kok MJ, et al. (1999) Can keratin 8 and 17 immunohistochemistry be of diagnostic value in cervical cytology? A feasibility study. Cancer 87: 87-92. [Crossref]
- Schmitt M1, Bravo IG, Snijders PJ, Gissmann L, Pawlita M, et al. (2006) Bead-based multiplex genotyping of human papillomaviruses. J Clin Microbiol 44: 504-512. [Crossref]
- Agorastos T, Koutkias V, Falelakis M, Lekka I, Mikos T, et al. (2009) Semantic integration of cervical cancer data repositories to facilitate multicenter association studies: the ASSIST approach. Cancer Inform 8: 31-44. [Crossref]
- Daud II, Scott ME (2008) Validation of reference genes in cervical cell samples from human papillomavirus-infected and -uninfected women for quantitative reverse transcription-PCR assays. Clin Vaccine Immunol 15: 1369-1373.
- Metzger C, Pittl A, Kaufmann AM, Agorastos T, Chatzistamatiou K, et al. (2016) A New Sandwich ELISA Test Simultaneously Detecting E7 proteins of HPV-16, 18, and 45. Clin Microbiol 5: 260.




