Poly(ADP-ribose) polymerase (Parp) inhibitor 3-aminobenzamide (3-AB) pretreatment suppresses G1 arrest and enhances G2 arrest after gamma-irradiation in mouse embryonic fibroblast C3D2F1 3T3-a cells. 3-AB partially inhibits Waf1/Cip1/p21 and Mdm2 mRNA inductions, which are transcriptionally activated by p53, suggesting that poly(ADP-ribosylation) is involved in the downstream of p53 dependent signal transduction after gamma-irradiation in C3D2F1 3T3-a cells. In this study we further examined the involvement of poly(ADP-ribosylation) in cell cycle arrest. Effect on G1 arrest suppression was lost when 3-AB was added 6 hrs after irradiation. When C3D2F1 3T3-a cells were synchronized by serum starvation, and gamma-irradiated, the peak time of DNA synthesis was not changed but the ratio of DNA synthesis was decreased in gamma-irradiated cells, where 3-AB pretreatment slightly enhanced the decrease of this ratio. 3-AB decreased G1 arrest in mouse embryonic fibroblast Swiss3T3 cells dose-independent manner whereas G1 arrest was not affected at low doses in FM3A and NRF49F cells. To confirm the inhibitory effect of 3-AB on Parp activity, NAD level change was measured after gamma-irradiation. We observed NAD decrease induced by gamma-irradiation was prevented by the 4 mM 3-AB, suggesting sufficient inhibition of cellular Parp activity at 4 mM concentration. These results suggested that the effect of Parp inhibitor on G1 arrest after gamma-irradiation depends on cell phenotypes.
In the cell nuclei of higher eukaryotes such as mammals, poly(ADP-ribose) polymerase (Parp) 1 recognizes DNA strand breaks and immediately synthesizes poly(ADP-ribose) chains by using β-NAD as substrate. Parp1 is constitutively present in the nucleus at a ratio of approximately one per dozens of kilobases of DNA [1]. The observations that Parp1 is constitutively present in the nucleus, recognizes DNA strand breaks in a specific manner, and catalyzes a poly(ADP-ribose) chain synthesis reaction by using NAD suggest that it could be a candidate as a sensor for monitoring DNA damage. Indeed, previous studies have shown that when gamma irradiation was carried out in the presence of a Parp inhibitor, the cytotoxicity of the treatment was enhanced [2]. In addition, given that Parp1 specifically recognizes the free ends of a broken DNA molecule, Parp1 may have a functional role in cutting out and repairing the damaged regions of the DNA. Further, Parp1-mediated DNA repair in response to alkylating agents is inhibited in mutant cells with low expression of Parp1, as well as in dominant negative mutants overexpressing Parp1 DNA-binding site [3]. In experiments where anti-sense RNA against Parp1 inhibited its intracellular functions, the DNA repair process was delayed [4]. Findings in a cell-free DNA repair system showed that in the presence of NAD, the DNA repair process was transiently interrupted in a Parp-dependent manner [5]. Thus, Parp1 appears to be involved in DNA damage repair, especially where DNA strand breaks occur [1].
An inhibitor of Parp, 3-aminobenzamide (3-AB) was previously reported to partially inhibit Waf1/Cip1/p21 and Mdm2 mRNA induction, which are transcriptionally activated by p53 [6,7], suggesting that poly(ADP-ribosylation) is involved in the downstream of p53 dependent signal transduction after gamma-irradiation in C3D2F1 3T3-a cells [7]. In this study we further examined the involvement of poly(ADP-ribosylation) in cell cycle arrest using 3-AB as well as tannic acid, which is known to inhibit poly(ADP-ribose) glycohydrolase (Parg) [8,9].
Two methods are currently used to examine whether poly(ADP-ribose) synthesis is involved in DNA repair after DNA damage due to gamma irradiation. The first method involves the preparation of knockout mice with constitutively disrupted Parp1 gene function or transfection of DNA into cultured cells for conditional disruption of Parp1. The second method involves the use of specific inhibitors to inhibit Parp. However, the former method, when used for cells in culture, involves the use of DNA transfection, which, in itself, is predicted to cause damage to other genes in the cell. With respect to the latter method, the effect of Parp1 inhibitors on poly(ADP-ribose) synthesis needs to be confirmed. The poly(ADP-ribose) synthesis reaction consumes intracellular NAD as a substrate, and thus, quantification of intracellular NAD could be used as an indicator of the poly(ADP-ribose) synthesis reaction. Berger et al. previously reported that when L1210 cells were treated with the alkylating agent MNNG (2 µg/mL), the amount of intracellular NAD decreased to approximately 50% of its normal level after 30 minutes [10]. In addition, Shall et al. previously reported that when L1210 cells were irradiated with 100 Gy by using 60Co as a radiation source, the amount of intracellular NAD decreased to approximately 50% of its normal amount after 30 minutes [11]. Here in accordance with the method described by Santi et al., intracellular nucleotides were separated by high performance liquid chromatography (HPLC), and NAD level was also quantified [12].
Cell culture, gamma-irradiation, and flow cytometry
C3D2F1 3T3-a cell line, a fibroblast cell line that was established from 14-day-old embryos of C3H/HeJ and DBA/2J mice, was used in this study [13]. The cells were established and provided by Professor Katsuhiro Ogawa and his collaborators at Asahikawa Medical University. Cells were cultured with DMEM [13] (ICN Biochemical Inc. Costa Mesa, CA, USA) containing 10% fetal bovine serum (FBS) supplemented with 1% penicillin/streptomycin. The cells were passaged every 3 days. The doubling time was approximately 17 hours. This cell line has been reported to have no mutation in exon 5 to exon 9 of the p53 gene [5,13]. Swiss 3T3 [14] and FM3A clone 28 [15] and NRK49F [16] cells were also cultured with RPMI1640 medium containing 10% FBS supplemented with 1% penicillin/streptomycin. Cell synchronization was carried out by incubation of the cells with medium containing 0.25% FBS for 44 hrs and released cells by changing the medium containing 10% FBS and 4 hrs later, cells were treated with chemicals. DNA synthesis was measured by pulse-labeling with [3H]-thymidine (New England Nuclear) and trichloroacetic acid precipitation method. Cells were irradiated with a 60Co gamma-irradiator at 1 Gy/min. Cell cycle states were analyzed by pulsing cells with 10 µM bromodeoxyuridine (BrdU, Sigma) for 30 min at a selected period after gamma-irradiation. After irradiation, cells were harvested, fixed with 70% ethanol, denatured with 4 N hydrochloric acid, treated with RNase A, and stained with FITC-conjugated anti-BrdU antibody (Becton Dickinson, Mountain View, CA, USA) and with propidium iodide (Sigma), then analyzed by two-dimensional flow cytometry using FACScan (Beckton Dickinson).
Chemicals
3- Aminobenzamide (3-AB) was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Tannic acid was purchased from Wako Chemicals (Tokyo, Japan).
Quantification of intracellular NAD using HPLC
C3D2F1 3T3-a cells were irradiated with gamma rays two days after seeding. Cell pellets were harvested with PBS(-)[10], suspended in 100 mL of 0.3 M trichloroacetic acid, and placed on ice for 30 min while being vigorously stirred [11] at 1 min intervals. After centrifugation, the supernatants were stirred vigorously in equivalent amounts with 1,1,2-trichloro-1,2,2-trifluoroethane (Nacalai Tesque, Kyoto, Japan) containing 0.5 M tri-n-octylamine (Tokyo Chemical Industry Co., Ltd, Japan). The acid was then neutralized [12], and the acid-soluble fraction extract was adjusted to obtain a cell concentration of 3 × 106 cells per 100 mL.
The samples were separated by using a Partisil 10-SAX column, 4.6 mm × 250 mm, with 10-µm particle size (Sumitomo Analysis Center, Osaka, Japan). A low-concentration buffer (7 mM NH4H2PO4, pH 3.8) and a high-concentration buffer (250 mM NH4H2PO4, 500 mM KCl, pH 4.5) were used as eluting solutions. The samples were injected and were eluted in the low-concentration buffer for 6 min. Subsequently, the samples were eluted for 30 min in a gradient ranging from the low-concentration to high-concentration buffer. Following this gradient, the column was washed with high-concentration buffer for an additional 24 min. Flow velocity was 3 mL/minute and detection was carried out with a UV wavelength of 254 nm by using a TOYO SODA UV-8000. NAD was eluted with a retention time of 6 min. The UV absorption peak was analyzed by using a photodiode array, and the findings showed that it matched that of NAD. Therefore, the single peak was confirmed to be that of NAD. NADH and NADP were eluted with retention times of 13.3 and 17.9 min, respectively, and were distinct from NAD.
Decrease of G1 arrest suppression effect by delayed treatment of 3-AB
We previously showed that when 3-AB was added to C3D2F1 3T3-a cells 2 hrs before gamma-irradiation, abrogated G1 arrest and enhanced G2 arrest were observed 12 hrs after gamma-irradiation. As shown in Figure 1A, we analyzed the effect of addition timing of 4 mM 3-AB on G1 arrest suppression after gamma-irradiation at 2 Gy in C3D2F1 3T3-a cells. The effect of G1 arrest suppression was reduced with the delay in time of addition and when 3-AB was added 6 hrs after irradiation, the effect of G1 arrest suppression was almost completely lost.
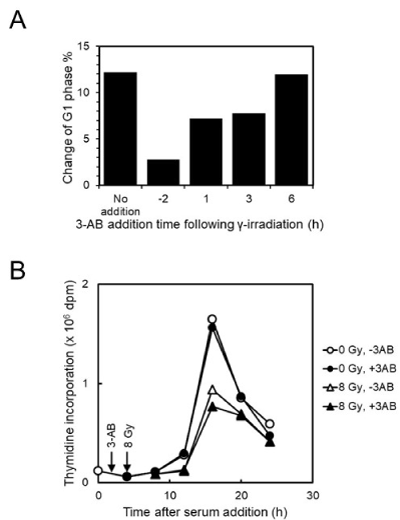
Figure 1. Effect of 3-AB on cell-cycle arrest and S-phase entry and progression suppression after gamma-irradiation in C3D2F1 3T3-a cells. A. Effect of addition timing of 4 mM 3-AB on G1 arrest suppression 12 hrs after gamma-irradiation at 2 Gy in C3D2F1 3T3-a cells. B. Effect of 2 hrs pretreatment of 4 mM 3-AB on DNA synthesis following gamma-irradiation at 8 Gy in C3D2F1 3T3-a cells synchronized by serum starvation
We further tested DNA synthesis timing by synchronization with serum starvation in C3D2F1 3T3-a cells after gamma-irradiation (Figure 1B). When synchronized cells were pretreated with 4 mM 3-AB for 2 hrs and irradiated at 8 Gy, the peak of DNA synthesis was observed at 16 hrs after release of serum starvation both in the mock-irradiated and irradiated cells in the absence or presence of 3-AB, whereas the ratio of DNA synthesis was decreased in gamma-irradiated cells, and 3-AB pretreatment slightly enhanced the decrease of DNA synthesis.
Effect of 3-AB on cell-cycle arrest in different cell lines
C3D2F1 3T3-a cells are known to possess functional p53-dependent signal transduction pathway. We examined the effect of cell types on cell-cycle arrest induced by gamma-irradiation (Figure2 and Table 1). When mouse embryonic fibroblast Swiss 3T3 cells were used, we observed that G1 arrest suppression 12 hrs after gamma-irradiation with 4 mM 3-AB pretreatment as in the case with C3D2F1 3T3-a cells. However, when mouse mammary-gland tumor derived FM3A and rat kidney-derived NRK49F cells were used, G1 arrest suppression 12 hrs after gamma-irradiation with 4 mM 3-AB pretreatment was not observed at lower dose of 1-2 Gy but could be observed at 4 Gy or higher doses. G2 arrest enhancement by 4 mM 3-AB pretreatment was observed at 2 Gy and higher doses in Swiss 3T3 and FM3A cells as in the case with C3D2F1 3T3-a cells and in NRK49F cells, G2 arrest enhancement by 4 mM 3-AB pretreatment was only observed at 4 Gy.
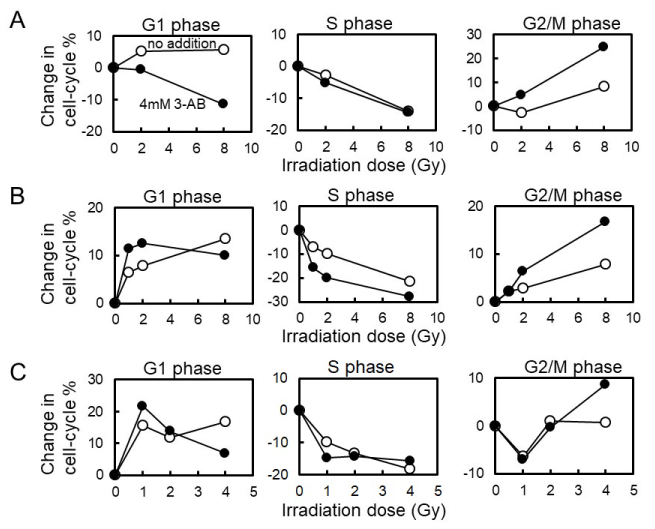
Figure 2. Effect of 3-AB on cell-cycle arrest 12hrs after gamma-irradiation in different cell lines. A, Swiss3T3 cells. B, FM3A cells. C, NRK49F cells
Table 1. Effect of 3-AB on cell-cycle arrest 12 hrs following gamma-irradiation in cell lines
|
|
Change in Cell-cycle Phase %*
|
|
|
G1
|
G2
|
|
Cell Line
|
-
|
3-AB (4 mM)
|
-
|
3-AB (4 mM)
|
|
C3D2F1 3T3-a
|
11.3
|
-6.2
|
10.0
|
13.0
|
|
Swiss 3T3
|
5.1
|
-0.4
|
-2.5
|
5.0
|
|
FM3A
|
8.2
|
12.8
|
3.1
|
6.6
|
|
NRK49F
|
12.1
|
13.9
|
1.1
|
-0.5
|
*(Cell-cycle phase % of gamma-irradiated cells) - (cell-cycle phase % of mock-irradiated cells)
Assessment of Parp activity inhibition by measurement of NAD level
To confirm the inhibitory effect of 3-AB on Parp activity, we measured NAD level change after gamma-irradiation in C3D2F1 3T3-a cells. The changes in the amounts of intracellular NAD after irradiation with gamma rays were measured in C3D2F1 3T3-a cells [17] as a preliminary study. Two days after seeding, the cells were irradiated with gamma rays at 10 Gy, 100 Gy, and 200 Gy. Thirty minutes later, NAD was quantified via HPLC. The results showed that with 10 Gy, there was no detectable change in intracellular NAD levels. However, the NAD levels decreased in a dose-dependent manner, with 100 Gy and 200 Gy causing a 40% and 60% drop in NAD, respectively (data not shown). From this study, it was suggested that to detect NAD decrease by consumption of NAD, which could be due to Parp-1 activity, 200 Gy irradiation was shown to be necessary. Figure 3A shows the separation pattern of the intracellular nucleotide pool in C3D2F1 3T3-a cells, as determined by HPLC.
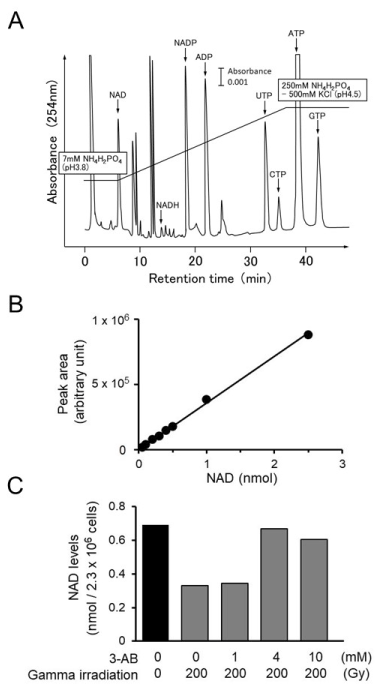
Figure 3. Measurement of intracellular NAD decrease in C3D2F1 3T3-a cells after gamma-irradiation. A. HPLC elution profile of intracellular nucleotides in C3D2F1 3T3-a cells. The vertical axis shows absorbance at 254 nm, and the horizontal axis shows retention time (min). Bar indicates 0.001 of absorbance. B. Calibration curve of NAD. The vertical axis shows area of peak (arbitrary unit), and the the horizontal axis shows amount of purified -NAD (nmol). C. Inhibitory effect of 3-AB on the decrease in intracellular NAD level 30 min after gamma irradiation. C3D2F1 3T3-a cells were irradiated with gamma irradiation at 200 Gy and NAD level was quantified. 3-AB was added at 1-10 mM
Therefore, in order to confirm whether 3-AB had an inhibitory effect on the intracellular synthesis of poly(ADP-ribose), 1 mM, 4 mM, and 10 mM 3-AB was added 1 h before irradiation with 200 Gy. The cells were collected 30 min after irradiation, and the intracellular NAD levels were quantified. In the absence of treatment, the intracellular NAD level was approximately 0.7 nmol/2.3 × 106 cells. The addition of 1 mM 3-AB suppressed the decrease in NAD levels caused by gamma irradiation by approximately 40%, and with 4 mM 3-AB, the decrease in NAD levels was suppressed by nearly 100% (Figur.
Effect of tannic acid on cell-cycle arrest following gamma-irradiation in C3D2F1 3T3-a cells
To evaluate the catabolic process of poly(ADP-ribose), we analyzed the effect of [8,9], on cell-cycle arrest following gamma-irradiation in C3D2F1 3T3-a cells. Tannic acid was added at 50 µg/ml to culture medium of C3D2F1 3T3-a cells 2 hrs before gamma-irradiation at 2 Gy and cells were incubated for 12 hrs. Before harvesting 12 hrs post-irradiation, cells were similarly pulse-labelled with BrdU for 30 min and analyzed with two-dimensional flow cytometry. Effect of different concentrations of tannic acid on cell-cycle arrest were further analyzed after gamma-irradiation at 2 Gy. As shown in Figures 4A and B, tannic acid did not cause suppression of G1 arrest nor the enhancement of G2 arrest in C3D2F1 3T3-a cells.
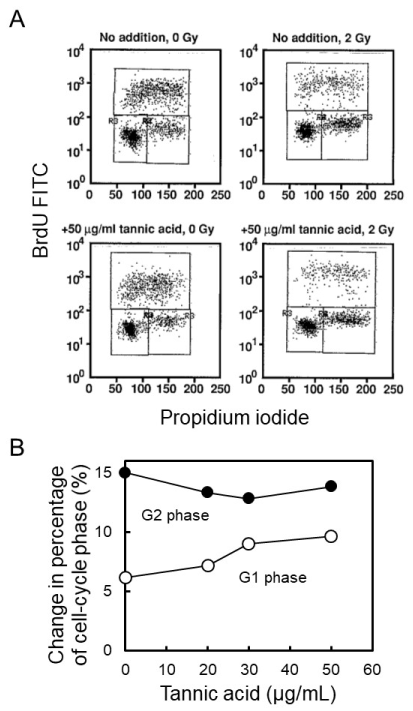
Figure 4. Effect of tannic acid on cell-cycle arrest following gamma-irradiation in C3D2F1 3T3-a cells. Tannic acid was added at 50 µg/mL to culture medium of C3D2F1 3T3-a cells 2 hrs before gamma-irradiation at 2 Gy and cells were incubated for 12 hrs. A. Before harvesting 12 hrs post-irradiation, cells were pulse-labelled with BrdU for 30 min and analyzed with two-dimensional flow cytometry. B. Effect of different concentration of tannic acid on cell-cycle arrest was analyzed after gamma-irradiation at 2 Gy
Poly(ADP-ribose) polymerase (Parp) inhibitor 3-AB pretreatment suppresses G1 arrest and enhances G2 arrest after gamma-irradiation in mouse embryonic fibroblast C3D2F1 3T3-a cells. 3-AB was previously shown to partially inhibit Waf1/Cip1/p21 and Mdm2 mRNA induction in several cell lines. Waf1/Cip1/p21 and Mdm2 mRNA are known to be transcriptionally activated by p53 protein. C3D2F1 3T3-a cells have been reported to harbor no mutation in exon 5 to exon 9 of the p53 gene and p53-dependent pathway seemed to be normal [13]. In this cell line, it has been suggested that poly(ADP-ribosylation) is involved in the downstream of p53-dependent signal transduction after gamma-irradiation [7]. To clarify the role of poly(ADP-ribosylation) in cell-cycle arrest after gamma-irradiation, we further analyzed the effect of 3-AB in C3D2F1 3T3-a cells as well as in other cell lines. In accordance with the predicted role of p53 in induction of Waf1/Cip1/p21 mRNA, which triggers the cell-cycle arrest, the effect on G1 arrest suppression was decreased when 3-AB was added 1-3 hrs after irradiation and the effect was completely lost when added 6 hrs after irradiation. It is notable that when C3D2F1 3T3-a cells were synchronized by serum starvation, and gamma-irradiated at 8 Gy, the peak time of DNA synthesis was not changed but the ratio of DNA synthesis was decreased in gamma-irradiated cells, where 3-AB pretreatment slightly enhanced the decrease of this ratio. In this limited experimental condition, the DNA damage is caused during the G0 phase and this could have affected the DNA damage type including single and double strand DNA breaks and affected the cell-cycle arrest profiles.
We also analyzed the effect of 3-AB in cell lines of different types. After gamma-irradiation, 3-AB decreased G1 arrest in mouse embryonic fibroblast Swiss3T3 cells dose-independent manner as in the case with whereas G1 arrest was not affected at low doses in FM3A and NRF49F cells. G2 arrest enhancement by 3-AB pretreatment was observed dose-independent manner in Swiss 3T3 and FM3A cells as in the case with G2 arrest enhancement by 4 mM 3-AB pretreatment was only observed not at 1-2 Gy but at 4 Gy.
To confirm the inhibitory effect of 3-AB on Parp activity, we also attempted to quantify intracellular NAD by HPLC. Prior to this, we used commercially purified β-NAD to establish a quantifiable range for NAD by using HPLC; this range was found to be between 0 and 2.5 nmol. Irradiation with gamma rays causes numerous DNA strand breaks in the cells. DNA strand breaks cause poly(ADP-ribose) synthesis to start immediately inside the nucleus. We verified whether poly(ADP-ribose) synthesis, which is supposed to be activated after irradiation with gamma rays, was inhibited by the Parp inhibitor, 3-AB. In our experiments, poly(ADP-ribose) synthesis was accompanied by the consumption of NAD; thus, we used HPLC to quantify intracellular NAD levels as an indicator of poly(ADP-ribose) synthesis activity. Our findings indicated that the decline in NAD levels after irradiation with 200 Gy was inhibited in a manner that was dependent on the concentration of 3-AB, and was nearly completely suppressed at a concentration of 4 mM 3-AB. Our study has shown that poly(ADP-ribose) synthesis reactions inside the cell were fully inhibited by 3-AB at a concentration of 4 mM. Conversely, the inhibitory effects of inhibitors on poly(ADP-ribose) synthesis were slightly suppressed by 10 mM 3-AB. The problem with using inhibitors is that it is difficult to determine if the inhibitor really inhibits the activity of the target enzyme inside the cell and whether the inhibitor inhibits the target enzyme exclusively. Therefore, it is desirable that the inhibitor fully suppresses the activity of the target molecule and can be used at concentrations as low as possible. HPLC allows for separating intracellular nucleotides; it also allows for determining the overall variations of intracellular nucleotides. Importantly, 4 mM 3-AB showed no marked influence on intracellular metabolism that was likely to affect intracellular nucleotides other than NAD. In our study, 4 mM was thus considered the optimal concentration for 3-AB.
To understand the impact of catabolism process of poly(ADP-ribose), we analyzed the effect of on cell-cycle arrest following gamma-irradiation in C3D2F1 3T3-a cells. poly(ADP-ribose) may not be critical in cell-cycle arrest after gamma-irradiation, although further studies with specific Parg inhibitors should be carried out.
In this study, we investigated the role of poly(ADP-ribosylation) after gamma-irradiation using immortalized cells, which retains basic p53-dependent signal-transduction pathway and in cancer cells. Taken together, the present results suggested that the effect of Parp inhibitor and the roles of poly(ADP-ribosylation) on G1 arrest after gamma-irradiation depends on cell phenotypes.
This work was supported in part by MEXT KAKENHI (15K14416) and Grant-in-Aid from Taijyu Life Social Welfare Foundation.
- Althaus FR, Richter C (1987) ADP-ribosylation of proteins. Enzymology and biological significance, Mol Biol Biochem Biophys 37: 1-237.
- Nduka N, Skidmore CJ, Shall S (1980) The enhancement of cytotoxicity of N-methyl-N-nitrosourea and of gamma-radiation by inhibitors of poly(ADP-ribose) polymerase. Eur J Biochem 105: 525-530. [Crossref]
- Molinete M, Vermeulen W, Burkle A, Menissier-de Murcia J, Kupper JH, et al. (1993) Overproduction of the poly(ADP-ribose) polymerase DNA-binding domain blocks alkylation-induced DNA repair synthesis in mammalian cells. EMBO J 12: 2109-2117.
- Ding R, Pommier Y, Kang VH, Smulson M (1992) Depletion of poly(ADP-ribose) polymerase by antisense RNA expression results in a delay in DNA strand break rejoining. J Biol Chem 267: 12804-12812.
- Satoh MS, Lindahl T (1992) Role of poly(ADP-ribose) formation in DNA repair. Nature 356: 356-358. [Crossref]
- Wieler S, Gagne JP, Vaziri H, Poirier GG, Benchimol S (2003) Poly(ADP-ribose) polymerase-1 is a positive regulator of the p53-mediated G1 arrest response following ionizing radiation. J Biol Chem 278: 18914-18921.
- Nozaki T, Masutani M (2018) Involvement of Parp1 in the downstream of p53 dependent signaling pathway induced after gamma-irradiation. J Transl Sci 5: 1-5.
- Sun Y, Zhang T, Wang B, Li H, Li P (2012) Tannic acid, an inhibitor of poly(ADP-ribose) glycohydrolase, sensitizes ovarian carcinoma cells to cisplatin. Anticancer Drugs 23: 979-990.
- Tikoo K, Sane MS, Gupta C (2011) Tannic acid ameliorates doxorubicin-induced cardiotoxicity and potentiates its anti-cancer activity: potential role of tannins in cancer chemotherapy. Toxicol Appl Pharmacol 251: 191-200.
- Berger NA (1985) Poly(ADP-ribose) in the cellular response to DNA damage. Radiat Res 101: 4-15. [Crossref]
- Goodwin PM, Lewis PJ, Davies MI, Skidmore CJ, Shall S (1978) The effect of gamma radiation and neocarzinostatin on NAD and ATP levels in mouse leukaemia cells. Biochim Biophys Acta 543: 576-582.
- Pogolotti AL Jr., Santi DV (1982) High-pressure liquid chromatography--ultraviolet analysis of intracellular nucleotides. Anal Biochem 126: 335-345.
- Tokumitsu M, Kadohama T, Ogawa K (1994) Infrequent loss of heterozygosity and mutation of the p53 gene in immortal and transformed mouse embryo fibroblasts. Mol Carcinog 10: 52-57.
- Mora PT, Fishman PH, Bassin RH, Brady RO, McFarland VW (1973) Transformation of Swiss 3T3 cells by murine sarcoma virus is followed by decrease in a glycolipid glycosyltransferase. Nat New Biol 245: 226-229.
- Takahashi M, Nishizawa M, Negishi K, Hanaoka F, Yamada MA, et al. (1988) Induction of mutation in mouse FM3A cells by N4-aminocytidine-mediated replicational errors. Mol Cell Biol 8: 347-352.
- Offringa R, Smits AM, Houweling A, Bos JL, van der Eb AJ (1988) Similar effects of adenovirus E1A and glucocorticoid hormones on the expression of the metalloprotease stromelysin. Nucleic Acids Res 16: 10973-10984.
- Benjamin RC, Gill DM (1980) ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J Biol Chem 255: 10493-10501. [Crossref]




