The 6-phospho-β-glucosidase can hydrolyze 6-phospho-β-glucoside to produce glucose and glucose 6-phosphate (G6P), both of which can enter the energy-generating glycolytic pathway. The 478-residue protein, 6-phospho-β-glucosidase BglA-1 from S. pneumoniae TIGR4 is encoded by the gene SP0303, which resides in an eight-gene operon composed by a lactose-type phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) which includes three separate subunits: EIIB (SP0305), EIIA (SP0308), EIIC (SP0310) and a multidomain transcriptional regulator. Here, through enzymatic analysis and sequence alignment, we present the substrate specificity of BglA-1. Glu375 and Glu176 function as the nucleophile group and the proton donor respectively. Three residues, Phe131, Tyr313 and Trp349 play important roles in the substrate specificity towards the β (1,4)-linked glucose 6-phosphate (G6P) bond. In addition, Ser432, Lys438 and Tyr440 participate in enzyme discrimination between the hydrolysis of phosphorylated and the non-phosphorylated substrates.
6-phospho-β-glucosidase, phosphoenolpyruvate (PEP)-dependent phosphotransferase system, substrate specificity
Streptococcus pneumoniae (S. pneumoniae), the important pathogenic microorganism with high mortality rates, is one of the leading causes of acute pneumonia, otitis media, meningitis, and many other serious diseases in humans [1-3]. Pneumococcal invasions lead to more than 1.6 million infections worldwide every year. In developed and developing countries, the treatment and prevention of diseases caused by S. pneumoniae still have great burden [4,5]. S. pneumoniae can colonize on the human nasopharyngeal mucosa when the baby is born just a few months. The microorganism need abundant energy sources, such as carton sources, to satisfy its growth in host cells during infection and colonization. There are 32 carbohydrates identified as energy sources in S. pneumoniae, including three-carbon molecules (glycerol), nine hexoses or hexose derivatives, three α-galactosides, two β-galactosides, four α-glucosides, seven β-glucosides, and six polysaccharides [6]. Carbohydrates, as the important carbon source of S. pneumoniae, its accumulation is facilitated by various sugar transporters, including phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) transporters, cation/proton-coupled transporters and ATP-binding cassette (ABC) transporters [7-17]. PEP-PTS systems which is the most ubiquitous transporters in bacteria, transport various carbohydrates, such as glucose, sucrose, mannitol, fructose, lactose, mannose, and cellobiose, the β-1,4 disaccharides of D-glucose [18]. The PEP-PTS systems are composed of three components: enzyme I (EI), heat-stable phosphocarrier protein (HPr), and enzyme II (EII) [13,19]. EI and HPr are nonspecific energy-coupling components. EII is consisting of three or four functional subunits (EIIA, EIIB, EIIC, and sometimes EIID) [20]. The transmembrane permeases, EIIC and EIID, which are responsible for recognition and binding of specific sugar substrates, transport sugars across the cell membrane into the cytoplasm. The sugar molecule is subsequently transferred to EIIA and EIIB domains and phosphorylated [10]. The PEP-PTS systems transport carbohydrates coinstantaneously with phosphorylation of C6 (11). The 6-phospho-β-glucosidase BglA-1 from S. pneumoniae belongs to glycoside hydrolase family 1 (GH-1) which possesses a varity of glycosyl hydrolases. Some 6-phospho-β-glucosidases are grouped in the glycosidase families GH-4, in addition to GH-1. GH-1 family belongs to a superfamily (or clan) termed GH-A, all members of which possess a catalytic domain of (α/β)8 TIM barrel fold. Members of GH-1, such as 6-phospho-β-glucosidase (EC 3.2.1.86), 6-phospho-β-galactosidase (EC 3.2.1.85) and β-glucosidase (EC 3.2.1.21) have different substrates specificity (see the CAZy database) [21]. The 6-phospho-β-glucosidase can hydrolyze 6-phospho-β-glucoside to produce glucose and glucose 6-phosphate (G6P), both of which can enter the energy-generating glycolytic pathway [22]. The 478-residue protein, 6-phospho-β-glucosidase BglA-1 from S. pneumoniae TIGR4 is encoded by the gene SP0303 which resides in an eight-gene operon (Figure 1A). The operon composed by a lactose-type PTS which includes three separate subunits: EIIB (SP0305), EIIA (SP0308), EIIC (SP0310) and a multidomain transcriptional regulator (Figure 1B) [23]. The primary sequence of BglA-1 is homologous to the following GH-1 enzymes of known structure: Lactococcus lactis PGALase, Bacillus polymyxa BglA, Human hCBG and S. pneumoniae BglA-2 [24-28]. The structure-based catalytic mechanism of GH-1 PGALase from Lactococcus lactis has been elucidated [24,25]. As reported previously,two conserved residues (glutamate or aspartate) function as catalytic residues, one as a nucleophile group and the other as a proton donor. The process of catalysis is as follow: the glycosidic oxygen of the substrate is protonated by the proton donor and forms the transient oxocarbenium state. Then nucleophilic residue attacks the protonated glycosidic bond, with the formation of a glycosyl-enzyme intermediate. Finally, the protonated glycosidic bond is broken by a proton provided by a water molecule, forming the product and returning the enzyme to its original protonated state [29,30].
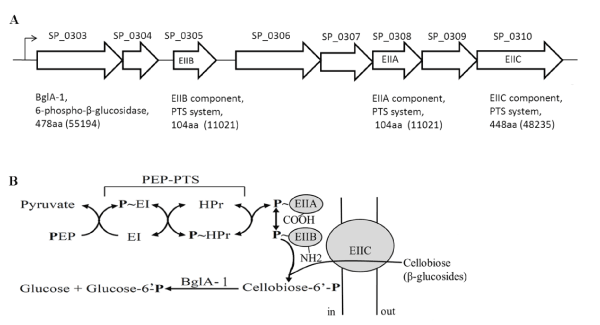
Figure 1A. genetic organization of the β-glucoside operon in S. pneumonia TIGR4. SP numbers represent the order of gene loci. Values in parentheses are calculated molecular weights of encoded proteins
B. translocation and phosphorylation of cellobiose by the multicomponent β-glucoside PEP-PTS and BglA-1-catalyzed hydrolysis of cellobiose-6´P
Previously, we have elucidated the mechanism of the substrate specificity towards the β (1,4)-linked glucose 6-phosphate (G6P) bond and the hydrolytic selectivity toward phosphorylated rather than non-phosphorylated compounds of 6-phospho-β-glucosidase BglA-2 from S. pneumoniae TIGR4 [28]. Here, through enzymatic analysis and sequence alignment, we present the substrate specificity of BglA-1 from S. pneumoniae TIGR4. Glu375 and Glu176 function as the nucleophile group and the proton donor respectively. Three residues, Phe131, Tyr313 and Trp349 play important roles in the substrate specificity towards the β (1,4)-linked glucose 6-phosphate (G6P) bond. In addition, Ser432, Lys438 and Tyr440 participate in enzyme discrimination between the hydrolysis of phosphorylated and the non-phosphorylated substrates.
Cloning and expression of BglA-1 and its mutants-The coding region of the bglA-1 gene was amplified from the genomic DNA of S. pneumoniae TIGR4. The bglA-1 gene and its mutants were respectively constructed into a pET28a-derived expression vector with N-terminal hexahistidine (6×His) tags. Both the wild-type and mutants were overexpressed in Escherichia coli (E. coli) strain Rosetta (DE3) (Novagen) using 2×YT culture medium (5 g of NaCl, 16 g of Bacto-Tryptone, and 10 g of yeast extract/liter). The transformed cells were grown at 37°C in 2×YT medium containing 30 μg/ml kanamycin and 34 μg/ml chloramphenicol. When the OD600nm reached about 0.6~0.8, the recombinant proteins were induced with 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 20 h at 16°C.
Purification of BglA-1 and its mutants—The cells were collected by centrifugation at 4000×g for 15 min and resuspended in 50 ml of lysis buffer (20 mM Tris-Cl, pH 7.5, 100 mM NaCl). After 5 min of sonication and 25 min of centrifugation at 12,000×g, the supernatant containing the target protein was collected and loaded onto a nickel-nitrilotriacetic acid column (GE Healthcare) equilibrated with the binding buffer (20 mM Tris-Cl, pH 7.5, 100 mM NaCl). The target protein was washed with the binding buffer (20 mM Tris-Cl, pH 7.5, 100 mM NaCl) and eluted with the same buffer containing 250 mM imidazole. Then the target protein was loaded onto a Superdex 200 column (GE Healthcare) equilibrated with the buffer containing (20 mM Tris-Cl, pH 7.5, 100 mM NaCl). Fractions containing the target protein were combined and collected. Samples for enzymatic activity assays were collected at the highest peak fractions without concentration. The purity of protein was assessed by SDS–PAGE, and the protein sample was stored at -80°C.
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the plasmid encoding the wild-type BglA-1 as the template. The mutant proteins were expressed, purified, and stored in the same manner as the wild-type protein.
Dynamic light scattering-Dynamic light scattering was measured through a DYNAPRO-99 (Wyatt Technology Corp, 6300 Hollister) using a 532 nm green laser. The sample was measured in single-use UV-plastic cuvettes (Wyatt Technology Corp, 6300 Hollister). Firstly, a time scale of the scattered light intensity fluctuations was measured. Secondly, the instrument was equilibrated for 2 min at 25°C. At last, the molecular weight of the solution protein BglA-1 was analyzed with the software Dynamic V6 (Wyatt Technology Corp, 6300 Hollister).
Enzymatic activity assays-The enzymatic parameters of wild-type BglA-1 and its mutants were measured with the substrate p-nitrophenyl-β-D-glucopyranoside-6-phosphate (pNpβGlc6P) following the previous processes with minor changes [31]. All of the reactions were performed at 37°C in the buffer containing 50 mM Na2HPO4 /50 mM NaH2PO4, pH 7.5 and triggered by the addition of BglA-1. Using a DU800 spectrophotometer (Beckman Coulter, Fullerton, CA), the change in absorption at 405 nm was monitored continuously. The reaction product PNP was calculated according to a standard curve of p-nitrophenol, which was described by Prag et al. The final Michaelis-Menten parameters (Vmax and Km) of BglA-1 were extracted from these data by nonlinear fitting to the Michaelis-Menten equation using the program Origin 7.5.
Preparation of 1-Phenyl-3-methyl-5-pyrazolone (PMP) derivatives of saccharides-PMP derivatization of saccharides was implemented as described previously with minor changes [32-34]. Briefly, the 10 μl reaction mixture was mixed with 10 μl of 0.3 M aqueous NaOH and a 10 μl 0.5 M methanol solution of PMP. The total reaction system of 30 μl mixture was placed to react for 30 min at 70°C, then cooled to room temperature, and neutralized with 10 μl of 0.3 M HCl. The creating solution was dissolved in 100 μl of chloroform. After vigorous shaking and centrifuging, the organic phase in the lower was carefully abandoned to remove the excess reagents. The extraction procedure was repeated three times. At last, the aqueous phase containing derivatives was diluted with 40 μl of water before HPLC analysis.
HPLC analysis-The assays towards specific substrate were performed at 37°C in a 10 μl system containing the buffer of 50 mM Na2HPO4/50 mM NaH2PO4, pH 7.5 and the disaccharide substrate cellobiose-6p´ with kinds of concentrations. The reactions were triggered by the addition of the purified enzymes and terminated by mixing with 10 μl of 0.3 M NaOH. After PMP derivation as mentioned above, the reaction product was centrifuged at 12,000×g for 10 min, and 15 μl supernatant was analyzed by HPLC system (Agilent 1200 Series). Glucose and G6P standards were quantified by HPLC analysis using different concentrations ranging from 0.1 to 1 mM. The mixing buffer composed of 20% acetonitrile and 100 mM Na2HPO4/NaH2PO4, pH 7.0 was processed as described previously for equilibration of the column (Eclipse XDB-C18 column, 4.6×150 mm; Agilent) and separation of the components at a flow rate of 1 ml/min [34]. Retention times of monosaccharides were determined by comparison with standard monosaccharide solutions. Kinetic parameters determinations through three independent experiments were made to calculate the means and standard deviations for the Km and kcat values.
Preparation of Cellobiose-6´P, Thiocellobiose-6´P, and pNPβGlc6P-Cellobiose was obtained from Pfanstiehl Laboratories, and thiocellobiose was purchased from Toronto Research Chemicals. Phosphorylation of the primary hydroxyl groups of the non-reducing glucose moiety in these O-β-linked disaccharides was as described previously [35]. In brief, phosphorylation was effected by incubation of the disaccharides with ATP-dependent β-glucoside kinase (BglK, EC 2.7.1.85) from Klebsiella pneumoniae. Phosphorylated derivatives were first isolated by Ba2+ and ethanol precipitation and further purified by ion exchange and paper chromatography. Structures and product purity were confirmed by thin layer chromatography, mass spectrometry, and NMR spectroscopy. Chromogenic pNPβGlc6P was prepared by phosphorylation of the C6 hydroxyl moiety of pNP-β-D-glucopyranoside with phosphorus oxychloride in trimethyl phosphate containing a small amount of water [34].
The state of BglA-1 in the solution-The plasmid of BglA-1 was constructed and overexpressed as described as the experimental procedures. The protein was purified though Ni-chelating affinity chromatography and size exclusion chromatography. And dynamic light scattering was carried out to confirm the state of BglA-1 in the solution. The results of size exclusion chromatography (Figure 2) and dynamic light scattering confirmed the existence of BglA-1 as a dimer in solution.
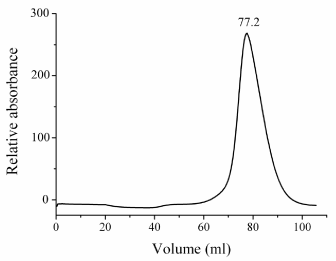
Figure 2. Gel-filtration chromatography of wild-type BglA-1 using Superdex 200 column. The chromogram represents the absorbance at 280 nm. The elution volume in miniliter of the peak was labeled on the top.
The enzymatic activity of BglA-1-The samples for enzymatic activity assays were collected at the highest peak fractions of size exclusion chromatography without concentration. The purity of protein was assessed by SDS–PAGE. Firstly, we tested enzymatic activity of BglA-1 for generic substrate (pNpβGlc6P) and specific substrate (cellobiose-6´p). The Km and kcat values for generic substrate were 654±4.8 μM and 136±6.4 s-1, respectively, and for specific substrate, 1924±8.9 μM and 152±3.8 s-1 (Table 1). These results suggested BglA-1 was a 6-phospho-β-glucosidase. Compared with other GH-1 members of known structures, the active-site residues of BglA-1 are highly conserved, especially two catalytic residues Glu176 and Glu375 (corresponding to Glu171 and Glu364 in BglA-2) (Figure 3). This suggests that BglA-1 presents a same catalytic mechanism as the previous known structures of GH-1 members [28,29]. The mutants E176A, E176Q, E375A and E375Q completely lost their activities towards pNpβGlc6P and cellobiose-6´p, suggesting the critical roles of Glu176 and Glu375 of BglA-1 in catalysis (Table 1).
Table 1. Kinetic constants of BglA-1 and mutants towards pNpβGlc6P and cellobiose-6´p
Substrate |
pNpβGlc6P |
Cellobiose-6´p |
Enzyme |
Km (μM) |
kcat (s-1) |
kcat/Km (s-1μM-1) |
Km (μM) |
kcat (s-1) |
kcat/Km (s-1μM-1) |
Wild Type |
654±4.8 |
136±6.4 |
2.1×10-1±4×10-2 |
1924±8.9 |
152±3.8 |
7.9×10-2±6×10-3 |
E375A |
ND |
ND |
ND |
ND |
ND |
ND |
E375Q |
ND |
ND |
ND |
ND |
ND |
ND |
F131A |
ND |
ND |
ND |
ND |
ND |
ND |
F131Y |
838±4.6 |
157±6.8 |
0.19±0.1 |
2102±12.3 |
129±9.8 |
6×10-2±4×10-3 |
Y313A |
ND |
ND |
ND |
ND |
ND |
ND |
Y313F |
ND |
ND |
ND |
ND |
ND |
ND |
W349A |
ND |
ND |
ND |
ND |
ND |
ND |
W349A |
ND |
ND |
ND |
ND |
ND |
ND |
Y440F |
ND |
ND |
ND |
ND |
ND |
ND |
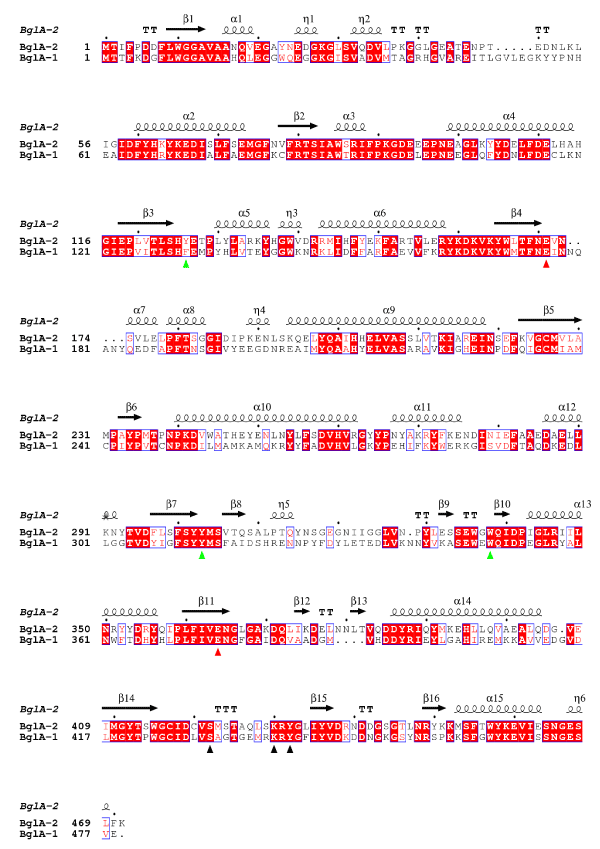
Figure 3. Sequence alignment of BglA-2 and BglA-1. Two conserved catalytic glutamate residues are marked with red triangle. The subsite +1 residues and the phosphate-binding residues are depicted by green and black triangles, respectively
We further tested the enzymatic activity of the wild-type and mutant proteins of BglA-1 for other substrates (gentiobiose-6´P or maltose-6´P) (Figure 4). No activity was showed towards these substrates. These results demonstrated the specificity of BglA-1 towards cellobiose-6´p.
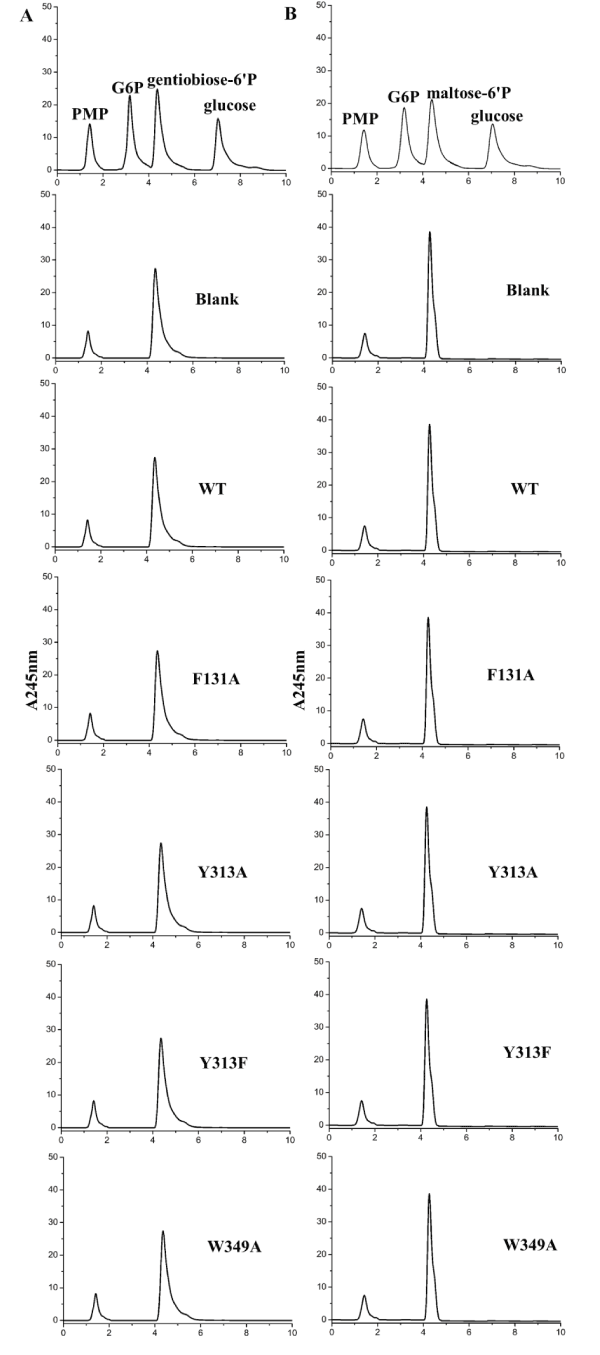
Figure 4. Enzymatic assays of wild-type BglA-1 and mutants towards gentiobiose-6'P (A) and maltose-6'P (B)
Key residues to the substrate specificity towards +1 site of cellobiose-6p´-Sequence alignment results revealed that three residues Phe131, Trp313 and Trp349 of BglA-1 (corresponding to Tyr126, Trp303 and Trp338 in BglA-2) were generally conserved (Fig. 3). To identify the putative roles of Phe131, Trp313 and Trp349 of BglA-1, enzymatic assays towards the site-directed mutants were carried out. The mutant F131A completely lost its activity whereas F131Y retained the activity of approximately 81% compared to the wild-type (Table 1). The results suggested that Tyr313 is indispensable for the activity by contributing to the hydrophobic pocket. In addition, the mutant W349A also absolutely lost its activity. These results clearly demonstrated that residues Phe131, Tyr313 and Trp349 play an important role in the substrate binding and thus might determine the specificity of +1 site sugar.
Key residues to the substrate specificity towards the phosphate group of cellobiose-6´p-As described previously, the phosphate-binding residues Ser424, Lys430 and Tyr432 in BglA-2 discriminated the phosphorylated from the non-phosphorylated sugar [34]. Compared between the sequences of BglA-1 and BglA-2, the residues Ser432, Lys438 and Tyr440 of BglA-1 were highly conserved, corresponding to Ser424, Lys430 and Tyr432 of BglA-2. Sequence analysis suggested that the three residues are conserved in GH-1 6-phospho-β-glucosidases from bacteria to protozoon as described previously [28]. As demonstrated previously, the residues were substituted by non-polar residues in other GH-1 glycosidases with non-phosphorylated substrate, such as β-glucosidase A from Bacillus polymyxa [26]. To verify the roles of the three residues, we tested the enzymatic activities towards phosphorylated substrate through a variety of mutants. The results indicate that S432A, K438A and Y440F completely abolished the activities, suggesting the key roles of Ser432, Lys438 and Tyr440 in determining the phosphorylated substrate (Table 1).
Sequence alignment indicated that two catalytic residues Glu176 and Glu375, the +1 site residues, Phe131, Trp313 and Trp349 and phosphate-binding residues, Ser432, Lys438 and Tyr440, were exclusively conserved. The results indicated that BglA-1 might adopt a similar overall structure and hydrolyze the similar substrate as other 6-phospho-β-glucosidase.
In summary, based on sequence alignment analysis and enzymatic assays, we not only elucidated the specificity towards +1 site glucose, but also the specificity towards -1 site phosphate group of 6-phospho-β-glucosides. The results expand our understanding on the substrate recognition pattern and specificity of GH-1 enzymes.
This work was supported by the Anhui Provincial Natural Science Foundation (Grant No. 1508085QC49), the school fund project of Anhui Medical University (Grant No. 2015XKJ031), the doctoral research fund project of the Second Affiliated Hospital of Anhui Medical University (Grant No.2014BKJ034) and Natural Science Foundation of Anhui Provincial Education Department (KJ2017A183).
The authors declare that they have no conflict of interest.
- Kadioglu A, Weiser JN, Paton JC, Andrew PW (2008) The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 6: 288-301. [Crossref]
- Cartwright K (2002) Pneumococcal disease in western Europe: burden of disease, antibiotic resistance and management. Eur J Pediatr 161: 188-195. [Crossref]
- Steinfort C, Wilson R, Mitchell T (1989) Effects of Streptococcus pneumoniae on human respiratory epithelium in vitro. Infect Immun 57: 2006-2013. [Crossref]
- Madhi SA, Levine OS, Cherian T (2008) Pneumococcal conjugate vaccine is efficacious and effective in reducing the burden of pneumonia. Bull World Health Organ 86: A-C. [Crossref]
- Rudan I, Boschi-Pinto C, Biloglav Z, Mulholland K, Campbell H (2008) Epidemiology and etiology of childhood pneumonia. Bull World Health Organ 86: 408-416. [Crossref]
- Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, et al. (2012) A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PloS one 7: e33320. [Crossref]
- Saier MH Jr1 (2000) Families of transmembrane sugar transport proteins. Mol Microbiol 35: 699-710. [Crossref]
- Ajdic D, Pham VT (2007) Global Transcriptional Analysis of Streptococcus mutans Sugar Transporters Using Microarrays. J Bacteriol 189: 5049-5059. [Crossref]
- Kundig W, Ghosh S, Roseman S (1964) phosphate bound to histidine in a protein as an intermediate in a novel phospho-transferase system. Proc Natl Acad Sci U S A 52: 1067-1074. [Crossref]
- Reizer J, Saier MH Jr, Deutscher J, Grenier F, Thompson J, et al. (1988) The phosphoenolpyruvate:sugar phosphotransferase system in gram-positive bacteria: properties, mechanism, and regulation. Crit Rev Microbiol 15: 297-338. [Crossref]
- Meadow ND, Fox DK, Roseman S (1990) The Bacterial PhosphoenolPyruvate: Glycose Phosphotransferase System. Annu Rev Biochem 59: 497-542. [Crossref]
- Postma PW, Lengeler JW, Jacobson GR (1993) Phosphoenolpyruvate:Carbohydrate Phosphotransferase Systems of Bacteria. Microbiol Rev 57: 543-594. [Crossref]
- Henderson PJ, Baldwin SA, Cairns MT, Charalambous BM, Dent HC, et al. (1992) Sugar-cation symport systems in bacteria. Int Rev Cytol 137: 149-208. [Crossref]
- Poolman B, Knol J, van der Does C, Henderson PJ, Liang WJ, et al. (1996) Cation and sugar selectivity determinants in a novel family of transport proteins. Mol Microbiol 19: 911-922. [Crossref]
- Davidson AL (2002) Mechanism of coupling of transport to hydrolysis in bacterial ATP-binding cassette transporters. J Bacteriol 184: 1225-1233. [Crossref]
- Davidson AL, Chen J (2004) ATP-binding cassette transporters in bacteria. Annu Rev Biochem 73: 241-268. [Crossref]
- Patzlaff JS, van der Heide T, Poolman B (2003) The ATP/substrate stoichiometry of the ATP-binding cassette (ABC) transporter OpuA. J Biol Chem 278: 29546-29551. [Crossref]
- Palmer RE, Anderson RL (1972) Cellobiose Metabolism in Aerobacter aerogenes. II. Phosphorylation of cellobiose with adenosine 5’-triphosphate by a b-glucoside kinase. J Biol Chem 247: 3415-3419.
- Cote CK, Cvitkovitch D, Bleiweis AS, Honeyman AL (2000) A novel beta-glucoside-specific PTS locus from Streptococcus mutans that is not inhibited by glucose. Microbiology 146: 1555-1563. [Crossref]
- Kempton JB, Withers SG (1992) Mechanism of Agrobacterium beta-glucosidase: kinetic studies. Biochemistry 31: 9961-9969. [Crossref]
- Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid. Biochem J 280: 309-316. [Crossref]
- Thompson J, Pikis A, Ruvinov SB, Henrissat B, Yamamoto H, et al. (1998) The gene glvA of Bacillus subtilis 168 encodes a metal-requiring, NAD(H)-dependent 6-phospho-alpha-glucosidase. Assignment to family 4 of the glycosylhydrolase superfamily. J Biol Chem 273: 27347-27356. [Crossref]
- Mckessar SJ, Hakenbeck R (2007) The two-component regulatory system TCS08 is involved in cellobiose metabolism of Streptococcus pneumoniae R6. Bacteriol 189: 1342-1350.
- Wiesmann C, Beste G, Hengstenberg W, Schulz GE (1995) The three-dimensional structure of 6-phospho-ß-galactosidase from Lactococcus lactis. Structure 3: 961-968. [Crossref]
- Wiesmann C, Hengstenberg W, Schulz GE (1997) Crystal structures and mechanism of 6-phospho-beta-galactosidase from Lactococcus lactis. J Mol Biol 269: 851-860. [Crossref]
- Sanz-Aparicio J, Hermoso JA, Marti'nez-Ripoli M, Lequerica JL, Polaina J (1998) Crystal structure of ß-glucosidase A from Bacillus polymyxa. Insights into the catalytic activity in family 1 glycosyl hydrolases. J Mol Biol 275: 491-502. [Crossref]
- Tribolo S, Berrin JG, Kroon PA, Czjzek M, Juge N (2007) The crystal structure of human cytosolic ß-glucosidase unravels the substrate a glycone specificity of a family 1 glycoside hydrolase. J Mol Biol 370: 964-975. [Crossref]
- Yu WL, Jiang YL, Pikis A, Cheng W, Bai XH, et al. (2003) Structural Insights into the Substrate Specificity of a 6-Phospho-ß-glucosidase BglA-2 from Streptococcus pneumonia TIGR4. J Biol Chem 288: 14949-14958. [Crossref]
- Koshland DE (1953) Stereochemistry and the mechanism of enzymic reactions. Biol Rev Camb Philos Soc 28: 416-436.
- Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon JP, et al. (1995) Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci USA 92: 7090-7094. [Crossref]
- Thompson J, Robrish SA, Bouma CL, Freedberg DI, Folk JE (1997) Phospho-beta-glucosidase from Fusobacterium mortiferum: purification, cloning, and inactivation by 6-phosphoglucono-delta-lactone. J Bacteriol 179: 1636-1645. [Crossref]
- Yang X, Zhao Y, Wang Q, Wang H, Mei Q (2005) Analysis of the monosaccharide components in Angelica polysaccharides by high performance liquid chromatography. Anal Sci 21: 1177-1180. [Crossref]
- Honda S, Akao E, Suzuki S, Okuda M, Kakehi K, et al. (1989) High-Performance Liquid-Chromatography of Reducing Carbohydrates as Strongly Ultraviolet-Absorbing and Electrochemically Sensitive 1-Phenyl-3-Methyl-5-Pyrazolone Derivatives. Anal Biochem 180: 351-357. [Crossref]
- Jiang YL, Yu WL, Zhang JW, Frolet C, Guilmi AD, et al. (2011) Structural Basis for the Substrate Specificity of a Novel beta-N-Acetylhexosaminidase StrH Protein from Streptococcus pneumoniae R6. J Biol Chem 286: 43004-43012. [Crossref]
- Thompson J, Lichtenthaler FW, Peters S, Pikis A (2002) ß-Glucoside kinase (BglK) from Klebsiella pneumoniae. Purification, properties, and preparative synthesis of 6-phospho-ß-D-glucosides. J Biol Chem 277: 34310-34321. [Crossref]




