Introduction: Several viruses are accused in the severe acute respiratory infections and their diagnosis profits from the assets of the multiplex PCR (polymerase chain reaction).
The aim of this study was to describe the general epidemiology of these infections in hospitalized children and adults and to compare the diagnostic performances of the multiplex to an uniplex PCR.
Methodology: Our study included 147 patients hospitalized for severe acute respiratory infections in the IBN SINA University Hospital Center in Rabat between October 2015 and August 2016. The samples were analyzed by multiplex PCR using FTD respiratory pathogens 21 plus and by uniplex PCR.
Results: We diagnosed a viral infection in 65% patients and the frequency was significantly higher in the pediatric population (73 vs 46%, p= 0,002). Influenza virus, respiratory syncytial virus (RSV), and human metapneumovirus showed a seasonal peak during the winter season, while enterovirus, adenovirus, parainfluenza virus 3 and rhinovirus do not have marked seasonal variation. Multiple viral infections were identified in 22% of positive cases and were detected only in the pediatric samples and exclusively for the age 0 to 2 years old (p=0.001). The comparison of the multiplex versus the uniplex technique found a perfect technical agreement for the detection of AH1N1 (k = 0.83) and satisfactory agreement for the detection of A / B VRS (k = 0.68).
Conclusion: we observed significant differences in viral infection rates and virus types among age groups and seasons and supported the contribution of the multiplex PCR in the diagnosis of severe acute respiratory infections.
Epidemiology, multiplex PCR, respiratory infection, viral infection
The acute respiratory infections remain the major cause of morbidity and mortality worldwide particularly among children, the elderly, the immunocompromised persons and those suffering of a subjacent lung or cardiac disease [1]. They represent as a consequence, a public health problem. Establishing a rapid etiological diagnosis of severe acute respiratory illness may improve treatment decisions and patient outcomes.
Worldwide, the viruses are the principal etiologic agents of these respiratory infections [2-4].
The major viral agents of acute respiratory infections include influenza A, B viruses (Flu), respiratory syncytial virus (RSV), human metapneumovirus (hMPV), parainfluenza (1-4) virus (PIV), adenovirus (ADV), rhinovirus, enterovirus , human coronavirus (HCoV), and the human bocavirus (HBoV) [5-8].
The clinical secondary symptoms with these various viruses can overlap and thus increase the risks of errors during the clinical diagnosis [9]. This requires a precise and fast laboratory diagnosis in order to improve the therapeutic decisions and the outcome of the patients.
Classically, the virological diagnosis of the respiratory infections rested on antigen detection and viral culture. However, the diversity of the implied viruses and the number of different viral strains make this diagnosis long and difficult [10].
In recent years, multiplex PCR methods have been developed, with the aim of detecting a large panel of respiratory pathogens in a single sample [11]. These techniques have been shown to be the least costly strategy, generating significant savings for hospitals [12]. It aids in the design of diagnostic strategies and can help to reduce unwarranted antimicrobial consumption [13-15].
Most of the available literature describing the epidemiology of respiratory viruses is focused on the pediatric population [16], a particular population [4], specific viral agents [10] or studies frequently limited to a fixed season. Two preliminary studies were conducted in Morocco and have already shown prevalence of the viruses in the respiratory infections at the child [17,18]. At the adult except for the flu virus, no other virus of the respiratory infections was studied, and the PCR multiplex is little used for the surveillance (supervision) of severe acute respiratory infections (SARI).
Our study aims to describe the general molecular epidemiology of viral respiratory infections in pediatric and adult hospitalized at Ibn Sina University hospital of Rabat, Morocco.
In addition we compare the diagnostic efficacies of a multiplex PCR assay (Kit FTD respiratory pathogens 21 plus) and an uniplex PCR.
Material
This study included 147 patients hospitalized for severe acute infection at Ibn Sina University Hospital, in the capital of Morocco between October 2015 and August 2016.
For each patient, a CRF was filled in. The data collected was: the age, sex, the ward, date of hospitalization, date of the symptom´s onset, the town of residence. The co-morbidities were also documented and concerned the medical antecedents (chronic cardiovascular diseases, lung diseases, diabetes, pregnancy, chronic respiratory insufficiency, hematologic diseases, and chronic neurological diseases). The vaccine statute against influenza of the patient at the time of the season autumn winter 2015-2016 with respect to the influenza was also indicated. In order to see whether the case fullfiled the WHO defination of clinically severe pneumonia, the precise clinical characteristics of the episode in progress were collected, including the existence of general signs (fever, asthenia, anorexia, hypotension, confusion, convulsion, myalgias, digestive signs), the existence of high respiratory signs (rhinitis, pharyngitis, dysphonia) and of low respiratory signs (breathing difficulty, cough, bronchial obstruction, wheezing, hearth ausculatoire) were also well informed: The lent of stay ,the nature of the sample obtained from the patient and the outcome of the patients (cured or deceased).
Further, specimens were considered ‘pediatric’ if they were from patients under 16 years of age, ‘adult’ if from patients more than 16.
Nasopharyngeal specimens carried out using the Copan CLASSIQSwabsTM and FLOQSwabsTM © provided by the laboratory.
The specimens were conveyed in a specific medium transport (universal transport medium [UTM-RT]) to Central Laboratory of virology where a Multiplex real-time PCR was carried out.
The data was collected anonymously.
Laboratory methods
The samples received at the central laboratory of virology were analyzed by the Multiplex real-time PCR (one step reverse transcription) by using Kit FTD Respiratory Pathogens 21 plus (Fast Track Diagnostics Luxembourg©) which allows the detection of 21 pathogens (viruses and bacteria).
It is about a qualitative test of search for viral nucleic acid allowing simultaneous detection and the identification of several respiratory viruses tropism starting from nasopharyngeal swabs and aspirates, bronchial and/or tracheal aspirates and bronchoalveolar lavage (BAL) specimens.
The FTD multiplex real-time RT-PCR assay consists of six separate primer/probe mixes: mix 1: influenza A virus (Inf A), rhinovirus (RV), influenza A H1N1 pdm09, influenza B virus (Inf B); mix 2: coronavirus (CoV) NL63, 229E, OC43 and HKU1, mix 3: parainfluenza virus (PIV) 2, 3 and 4 and internal control ; mix 4: PIV 1, human metapneumovirus (HMPV) A and B, Mycoplasma pneumonia and human bocavirus (HBoV); mix 5: respiratory syncytial virus (RSV), adenovirus (AdV), enterovirus (EV) and parechovirus (PeV); mix 6: Staphylococcus aureus, Chlamydophila pneumonia, Streptococcus pneumonia, Haemophilus influenza B. Individual assays within each pool are distinguished by the use of different probe fluorophores. Each kit also contains a positive plasmid control pool and detailed instructions on test performance. The FTD 21 plus assay was performed following the manufacturer’s recommendations.
An RNA EAV (Equine arteritis virus) serves as an internal extraction control when spiked into the sample and into the negative control during extraction.
DNA or RNA was extracted from 190 µl of samples and eluted into 60 µl, using the QIAamp viral RNA Minikit Virus Spin kit (Qiagen)
Briefly, 150 µL of 2×RT-PCR buffer was combined with 18,75 µL of each primer/probe pool and 12 µL of 25× enzyme mix and 15 µL of each mixture was then added to 12 wells of a PCR plate (10 sample reactions plus one positive and one negative virus control). Ten µL of sample DNA extract or controls were then added to the respective wells of each primer/probe pool. The following cycling conditions were performed on a thermocycler CFX96 of Biorad: 15 min at 42°C, 3 min at 94°C and 45 cycles of 8 s at 94°C and 34 s at 60°C. A positive test result was considered a well-defined curve that crossed the threshold cycle within 40 cycles. One positive control for viral and bacterial multiplex PCR reactions and a negative control tube are provided in the kit.
As for the PCR simplex, it was carried out at the National Laboratory of Hygiene by using a one step reverse transcription (Super Script III Platinum One step q Rt-PCR kit). The viral RNA was extracted from the sample on the iPrep Purelink Virus Kit (ThermoFisher Scientific, Carlsbad, CA the 92008 USA). The specific probes and primers were provided by the CDC. The following cycling conditions were performed on a 7500 Fast 96-well (Applied Biosystems, Foster City, CA, the USA) according to the protocol of the CDC (US centers for Disease Control).
Statistical analysis
Statistical analysis was performed using SPSS 13.0 (SPSS Inc., Chicago, IL, USA). Quantitative variables were expressed as mean ± SD or medians and interquartile range, depending on their distribution. For qualitative variables, data were presented as number and proportion and were compared using either the Chi-square test or the Fisher exact test. The degree of agreement between simplex and multiplex technical was determined by the test kappa of Cohen. A p-value of <0.05 was considered statistically significant.
This prospective study enrolled a continuous series of 147 patients hospitalized for severe acute infection at Ibn Sina University Hospital, in the capital of Morocco between October 2015 and August 2016.
We collected 94 specimens from males and 53 from females. The number of children (age ≤16 years) was 103/147 (70%) and their median age was 12 [6-22] months, (range: 0 month-14 years). The age bracket the most represented was 0-2 years with 83 (84%). The number of adults was 44/147 (30%) and their mean age was 49 ± 18 years (range: 17-94).
90% of the patients fulfilled the WHO definition of clinically severe pneumonia [19] including fever or antecedents of fever ≥38°C, cough, beginning of the disease in the last ten days and requiring an hospitalization.
None of the patients included in the study was vaccinated against influenza.
The median of hospitalization duration was 4 [2-11] days. On the whole 84 patients (57%) were hospitalized in the infectious pediatry service, 19 (13%) in the pediatric intensive care unit, 37 (25%) in the adult medical intensive care and seven (5%) in various adult medical departments.
Most of the samples studied were nasopharyngeal swabs, 96% (n= 116). For some patients, bronchoalveolar lavages, 1% (n= 1), bronchial aspiration 1% (n= 1) or PDP 2% (n=3) were obtained.
At least one viral respiratory pathogen was detected by multiplex PCR in 95 of the 147 (65%) specimens analyzed. 34 (23%) specimens were negative and in 18 (12%) specimens we detected a pure bacterial infection without viral coinfection. Multiple infections were observed in 41 samples (37 with two pathogens and four with three pathogens).
The overall positivity rate (PR) for any respiratory virus was 72% in children and 45% in adults.
Thus, on the whole, 122 viruses and 78 bacteria were detected: 34 (28%) cases of rhinovirus infections, 21 (17%) cases flu infections including 16 influenza A(H1N1), 2 influenza A not H1N1 and 3 influenza B, 15 (12 %) cases of RSV A/B, 14 (11%) cases of human metapneumovirus A/B, 11 (9%) cases parainfluenza virus including eight parainfluenza 3, two parainfluenza 4, one parainfluenza 1, nine (7%) cases of enterovirus, seven (6%) cases of adenovirus, seven (6%) cases of coronavirus including two coronavirus 229E, two coronavirus HKU, two coronavirus OC43 and one coronavirus NL63, four (4%) cases of bocavirus. Figure 1 describes the respective prevalence of different types of viruses according to age group and services.
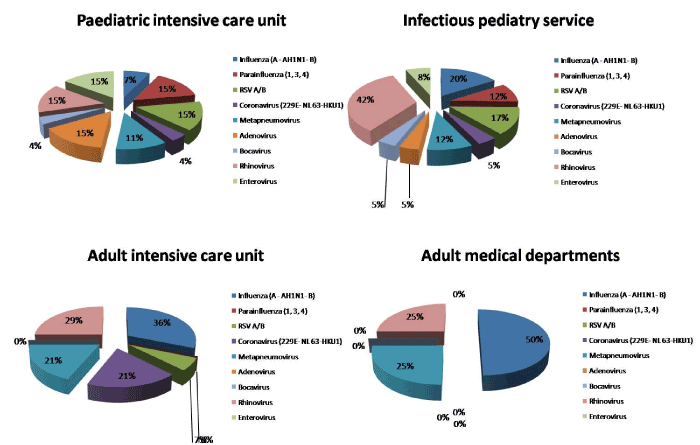
Figure 1. Prevalence of the different types of viruses according to age group and ward in admitted patient at University hospital of Rabat for acute severe respiratory infection.
Multiple viral infections, (defined as the simultaneous detection of more than one virus in one clinical specimen), were apparent in 23 samples (21 were positive for two viruses and two for three viruses). The most frequent co-infections were enterovirus with adenovirus (13%) and rhinovirus with RSV (3%).
With regard to the seasonal distribution of the 122 viruses, a peak was observed in winter: 62% (76/122). In spring, we found 15% (18/122) of viral infections, in summer 15% (18/122) and autumn 8% (10/122).
Regarding the distribution of respiratory viruses over the year, influenza virus, RSV and human metapneumovirus showed a clear seasonal pattern, with a marked increase in positivity rate throughout the winter months, while enterovirus, adenovirus, rhinovirus and parainfluenza 3 prevalence remained relatively stable over the year and did not have marked seasonal variation. Seasonal prevalence patterns are depicted in Figure 2.
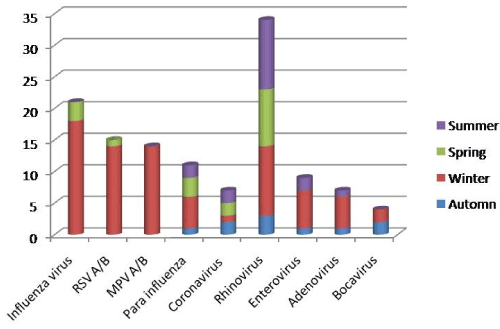
Figure 2. Prevalence of the different types of viruses according to age group and ward in admitted patient at University hospital of Rabat for acute severe respiratory infection.
We noted 107 (92%) cases of cure and nine (8%) cases of death during this study.
Among the nine cases of death, five presented a viral infection. Two were children hospitalized in pediatric intensive care from whom we isolated, for one, an enterovirus and an adenovirus, and for the other, a human metapneumovirus. The tree other cases of death corresponded to adults hospitalized in adults intensive care among them were respectively isolated an influenza AH1N1, an influenza A not H1N1 and a coronavirus 229E.
Analytical results
The highest positivity rate 73% was found in respiratory samples collected from pediatric patients, versus 46% in respiratory samples from adult patients. Pediatric patients had a significantly higher positivity rate (p = 0.002).
The correlation between the presence of the viruses and age group, objectified statistically significant differences with regard to the RSV and the parainfluenza: indeed we found 14 cases of RSV (93%) at the pediatric patients and only one case (7%) in adult patients (p=0.04). These 14 cases related to only the age range 0-2 years. Also, the 11 cases of parainfluenza virus were isolated only in the pediatric patients (p=0.034) and were distributed on the various age brackets.
Multiple viral infections were only diagnosed among pediatric patients and exclusively the age range 0-2 years. No multiple viral infections was noted at the adult patients (p= 0.001).
We also compared the viral distribution according to the seasons and we belonged to the statistically significant differences for the RSV and the metapneumovirus: indeed, the infections with RSV were diagnosed between mid-January and at the end of April with a peak in February: 13 cases detected in winter and one case in spring (p= 0.008). The human metapneumovirus circulated exclusively in winter (p=0.002).
Comparative study PCR multiplex/uniplex
We assessed the correlation between the results obtained by multiplex real-time PCR and by uniplex PCR (taken as reference) for the detection of influenza virus. On 118 samples: 108 cases were negative and 10 positive on the two techniques. We raised one case of false-negative and two cases of false-positives. It results from this a sensitivity of 89% and a specificity of 98% of the multiplex real-time PCR compared to the uniplex. We founded a k= 0,83 testifying an almost perfect agreement between the two techniques with regard to the detection of influenza AH1N1.
We also compared the two techniques for the detection of VRS A/B on 53 samples: 45 cases were negative and eight positive. We raised four cases of false-negatives and one case of false-positive. It results from this a sensitivity of 64% and a specificity of 98% for detection of the VRS by the multiplex real-time PCR compared to the uniplex. The k= 0,681 testifying substantial agreement between the two techniques for the detection of RSV A/B.
We described the epidemiological patterns of respiratory viruses among hospitalized patients of all ages screened at a University Hospital for a 10 months period. We diagnosed a viral infection in 65% and it was significantly higher in the pediatric population (73 vs 46%, p= 0,002). Influenza virus, VRS, and human metapneumovirus showed a seasonal peak during the winter season, while enterovirus, adenovirus, parainfluenza 3 and rhinovirus do not have marked seasonal variation. Multiple viral infections were identified in 22% of positive cases and were detected only in the pediatric samples and exclusively for the age bracket 0-2 years old (p=0.001).
Most of the samples studied were nasopharyngeal swabs. Indeed according to the studies, the viral bronchopulmonary infection can be emphasized by sample not very invasive specimen carried out in an easily accessible site in current practice [16].
The introduction of multiplex PCR has revolutionized the management of community-acquired acute respiratory illness; the increased sensitivity of molecular diagnostic methods provides a more faithful depiction of the epidemiology of respiratory viruses at the ‘front line’ [20,21]. Thus, we studied the viral epidemiology of these patients by means of a molecular analysis (PCR) viral multiplex, by comparing the distribution of the infections and the viruses according to the age, the service and the seasons.
Overall, the positivity rate (PR) for any respiratory virus was 65% and it was significantly higher in the pediatric population (72%). This could partially be explained by the viral load in children which is significantly higher than in adults. Our results are comparable to other studies that used swabs and multiplex real time PCR. Bouvet et al. reported a PR of 61% [1], Ambrosioni and al. reported a PR of 43.2% and a rate of 80% in children [4]. A study conducted in Morocco, four years ago, on the same population but only pediatric showed a PR of 92% [17].
Influenza viruses and rhinovirus had the highest prevalence in our study at any age confused. The RSV and the parainfluenza viruses were found primarily in children. Moreover the RSV was insulated only in infants. The human metapneumovirus was insulated as well in children as in adults. In the study of Ambrosini and al. [4] following pediatric and adults patients the most prevalent virus was picornaviruses. The study of Jroundi and al. conducted in Morocco, four years ago, only in pediatric population, detected rhinovirus and RSV more frequently [17].
The analysis of the seasonal variation of the isolated viruses in the run of this study confirmed the seasonal character of the influenza viruses and the RSV which were detected starting from January until the end of April with a peak in February, and the human metapneumovirus which circulated exclusively in winter. Whereas the enterovirus, adenovirus, parainfluenza 3 and rhinovirus do not have marked seasonal variation.
Seasonality is well described for several viral respiratory pathogens. Existing evidence suggests that the seasonality of some pathogens may be driven by enhanced wintertime survival, and also by increased host susceptibility resulting from relative ‘wintertime immune suppression’ [22]. A cold environment decreases innate defense mechanisms such as mucociliary clearance, so increasing susceptibility to viral infections [23]. For those viruses showing seasonal patterns, the months in which the peaks occur may be determined by local topography and latitude, making comparisons among studies performed in different countries difficult [24]. However the study of Ambrosioni and al. which was held in Geneva, over one year, showed that the picornaviruses had the highest prevalence over all seasons although other viruses such as influenza virus and RSV increased their prevalence in winter [4].
In our study, multiple viral infections (MVI) were observed in 22% of positive cases overall. They were detected only in pediatric samples and exclusively for the age range 0-2 years. Other studies have shown different rates of co-infections. The study of Jroundi and al. conducted in the same hospital reported more MVI with a rate of 40, 4% in children between 2 to 59 months, co-infections with more than two virus were relatively uncommon. While two studies conducted in industrialized countries shown low rates. Indeed, Ambrosioni and al. reported an MVI rate of 12 % and were significantly more frequent in children (p<0.001) [4] and for Bouvet and al. [1] , MVI were found in 15% of cases.
The techniques of multiplex PCR, makes it possible to highlight co-infections which are not detected by the cellular culture [25]. Indeed, the multiplication of a virus can inhibit another virus and a weak viral inoculum of each stock can lead to a wrongfully negative result [26]. So the sensitivity of the PCR is quite higher than the cellular culture. Indeed the PCR makes it possible to identify more of viral respiratory infections compared to the culture [20, 21]. Thanks to this sensitivity, molecular biology can also be an epidemiologic tool to analyze the circulation of the viruses in the general population. However, because of this great sensitivity of the PCR, some discussed the value of a positive PCR. The capacity to detect the non replicative viruses or present of small quantity can be also a disadvantage. Indeed, the viruses detected by culture are replicative and infectious; there are thus few doubts about their implication. The immunofluorescence (IF) brings the proof of the attack of respiratory fabric by the description of viral inclusions in the infected cells. Moreover, this technique allows a semi quantitative evaluation. It is on the other hand more difficult to affirm the pathogenic role of the viruses detected by PCR. It detects the presence of the genome of the viruses in specimen, even only of fragments of this genome, but is not informative as for its replication and with its pathogenic role. However, Liolios and al. [27] recall that this pathogenic implication is ensured by the respiratory clinical signs having justified the realization of the sample. Other studies showed that remotely acute episode (more than one month) the PCR are negative. Thus, the detection of virus respiratory genome by PCR is not very frequent at the asymptomatic subjects. The detection of the ARN of rhinovirus is lower than, 5% in asymptomatic children [28]; and of the recent data obtained within population of adults that the frequency of detection of influenza virus or the RSV by PCR at asymptomatic subjects in periods of influenza epidemics or RSV is weak [29]. In addition, a fast etiologic diagnosis of the respiratory infections makes it possible to decrease the duration of hospitalization, the regulation of antibiotics or complementary examinations [30,31]. Lastly, the relative complexity of the technique is compared with culture which imposes a logistics and competences booked on specialized laboratories. The culture remains however an essential technique for the epidemiologic monitoring allowing the development of the composition of the influenza vaccines in the following years.
The cost of molecular technique is usually higher than classical techniques but the multiplex PCR allows the search for several viruses by the same technique, it is thus cheaper [32].
The comparison of the sensitivity and specificity of the PCR multiplex FTD 21 plus compared to the simplex PCR as reference (CDC protocol) found a sensitivity and a specificity of 89% and 98% respectively for detection of influenza virus and a perfect agreement of the two techniques. These data confirm the results of many studies showing a quite higher sensitivity of the multiplex PCR compared to the classical techniques for the influenza virus and a perfect agreement with the uniplex PCR [10,21,32,33].
Otherwise, the sensitivity and the specificity for detection of the RSV were respectively of 64% and 98% confirming results of other studies which brought back a sensitivity decreased of FTD 21 plus for detection of the RSV [34].
This kit marked IVD, include 6 multiplex which could be adapted to the epidemiologic context of the country. Except the price which could also be regarded as a disadvantage, this kit presents the disadvantage to detect viruses or bacteria with the state of bearing. In addition, it is difficult to modify in the event of emergence of a new virus of medical interest (MERS coronavirus) and it does not have software of interpretation of the results what makes the reading sometimes difficult.
The multiplex PCR, certainly expensive but effective, fast and powerful, deserves its use as a matter of routine because it allows a diagnosis even faster with, for the influenza and RSV, a sensitivity equivalent to that of the uniplex PCR. It aids in the design of diagnostic strategies and can help to reduce unwarranted antimicrobial consumption. It also allows the diagnosis of the multiple viral infections.
- Juven T, Mertsola J, Waris M, Leinonen M, Meurman O, et al. (2000) Etiology of community-acquired pneumonia in 254 hospitalized children. Pediatr Infect Dis J 19: 293-298. [Crossref]
- Henrickson KJ, Hoover S, Kehl KS, Hua W (2004) National disease burden of respiratory viruses detected in children by polymerase chain reaction. Pediatr Infect Dis 23: S11-S18. [Crossref]
- Ambrosioni J, Bridevaux PO, Wagner G, Mamin A, Kaiser L (2014) Epidemiology of viral respiratory infections in a tertiary care centre in the era of molecular diagnosis, Geneva, Switzerland, 2011-2012. Clin Microbiol Infect 20: O578-O584. [Crossref]
- Segondy M (2012) Les « nouveaux » virus respiratoires et leur diagnostic. Médecine thérapeutique / Pédiatrie 15: 123-132.
- Freymouth F, Vabret A, Legrand L, Eterradossi N, Lafay-Delaire F, et al. (2003) Presence of the new human metapneumovirus in French children with bronchiolitis. Pediatr Infect Dis 22: 92-94. [Crossref]
- Vabret A, Dina J, Gouarin S, Petitjean J, Corbet S, et al. (2006) Detection of the new human coronavirus HKU1: a report of 6 cases. Clin Infect Dis 42: 634-639. [Crossref]
- Zhou JY, Peng Y, Peng XY, Gao HC, Sun YP, et al. (2017) Human bocavirus and human metapneumovirus in hospitalized children with lower respiratory tract illness in Changsha, China. Influenza Other Respir Viruses 12: 279-286. [Crossref]
- Schnepf N, Resche-Rigon M, Chaillon A, Scemla A, Gras G, et al. (2011) High burden of non-influenza viruses in influenza-like illness in the early weeks of H1N1v epidemic in France. PloS one 6: e23514. [Crossref]
- Plouzeau C, Paccalin M, Beby-Defaux A, Giraudeau G, Godet C, et al. (2007) Diagnostic et surveillance épidémiologique des infections grippales et à virus respiratoire syncytial: intérêt de la PCR multiplex. Médecine et Maladies Infectieuses 37: 728-733.
- Mahony JB (2008) Detection of respiratory viruses by molecular methods. Clin Microbiol Rev 21: 716-747. [Crossref]
- Mahony JB, Blackhouse G, Babwah J, Smieja M, Buracond S, et al. (2009) Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J Clin Microbiol 47: 2812-2817. [Crossref]
- Leruez-Ville M (2007) [Diagnosis of viral respiratory infections]. Arch Pediatr 14: 404-409. [Crossref]
- Freymuth F, Vabret A, Petitjean J, Campet M (2000) Diagnostic des deux principales viroses respiratoires épidémiques : la grippe et les infections à virus respiratoire syncytial. Place de la virologie moléculaire. Médecine et Maladies Infectieuses 30: 191-201.
- Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE (2000) Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev 13: 559-570. [Crossref]
- Koenig-Zores C, Toll-Keller F, Ammouche C, Donato L (2013) L’analyse virologique des aspirations nasopharyngées reflète-t-elle l’infection respiratoire basse chez l’enfant ? Étude en PCR multiplex. Revue française d’allergologie 53: 59–64.
- Jroundi I, Mahraoui C, Benmessaoud R, et al. (2014) The epidemiology and aetiology of infections in children admitted with clinical severe pneumonia to a university hospital in Rabat, Morocco. J Trop Pediatr 60: 270-278. [Crossref]
- Jroundi I, Mahraoui C, Benmessaoud R, Moraleda C, Benjelloun B, et al. (2015) Knowledge gaps on paediatric respiratory infections in Morocco, Northern Africa. Arch Public Health 73: 28. [Crossref]
- WHO (2014) WHO Global Epidemiological Surveillance Standards for Influenza: Available: http://www.who.int/influenza/resources/documents/influenza_surveillance_manual/en/; 2012 [cited January 2014].
- Weinberg GA, Erdman DD, Edwards KM, Hall CB, Walker FJ, et al. (2004) Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. J Infect Dis 189: 706-710. [Crossref]
- Carrat F, Sahler C, Rogez S, Leruez-Ville M, Freymuth F, et al. (2002) Influenza burden of illness: estimates from a national prospective survey of household contacts in France. Arch Intern Med 162: 1842-1848. [Crossref]
- Fisman D (2012) Seasonality of viral infections: mechanisms and unknowns. Clin Microbiol Infect 18: 946-954. [Crossref]
- Giesbrecht GG (1995) The respiratory system in a cold environment. Aviat Space Environ Med 66: 890-902. [Crossref]
- Bloom-Feshbach K, Alonso WJ, Charu V, Tamerius J, Simonsen L, et al. (2013) Latitudinal variations in seasonal activity of influenza and respiratory syncytial virus (RSV): a global comparative review. PloS one 8: e54445. [Crossref]
- Grondahl B, Puppe W, Hoppe A, Kühne I, Weigl JA, et al. (1999) Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J Clin Microbiol 37: 1-7. [Crossref]
- Waner JL (1994) Mixed viral infections: detection and management. Clin Microbiol Rev 7: 143-151. [Crossref]
- Liolios L, Jenney A, Spelman D, Kotsimbos T, Catton M, et al. (2001) Comparison of a multiplex reverse transcription-PCR-enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J Clin Microbiol 39: 2779-2783. [Crossref]
- Nokso-Koivisto J, Kinnari TJ, Lindahl P, Hovi T, Pitkäranta A (2002) Human picornavirus and coronavirus RNA in nasopharynx of children without concurrent respiratory symptoms. J Med Virol 66: 417-420. [Crossref]
- Falsey AR, Criddle MC, Walsh EE (2006) Detection of respiratory syncytial virus and human metapneumovirus by reverse transcription polymerase chain reaction in adults with and without respiratory illness. J Clin Virol 35: 46-50. [Crossref]
- Adcock PM, Stout GG, Hauck MA, Marshall GS (1997) Effect of rapid viral diagnosis on the management of children hospitalized with lower respiratory tract infection. Pediatr Infect Dis J 16: 842-6. [Crossref]
- Woo PC, Chiu SS, Seto WH, Peiris M (1997) Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J Clin Microbiol 35: 1579-1581. [Crossref]
- Vabret A, Sapin G, Lezin B, Mosnier A, Cohen J, et al. (2000) Comparison of three non-nested RT-PCR for the detection of influenza A viruses. J Clin Virol 17: 167-175. [Crossref]
- Sakthivel SK, Whitaker B, Lu X, et al. (2012) Comparison of fast-track diagnostics respiratory pathogens multiplex real-time RT-PCR assay with in-house singleplex assays for comprehensive detection of human respiratory viruses. J Virol Methods 185: 259-266. [Crossref]
- Anderson TP, Werno AM, Barratt K, Mahagamasekera P, Murdoch DR, et al. (2013) Comparison of four multiplex PCR assays for the detection of viral pathogens in respiratory specimens. J Virol Methods 191: 118-121. [Crossref]
- Bouvet D, Gaudy-Graffin C, Garot D, Sunder S, De Gialluly C, et al. (2015) Diagnosis of community-acquired acute respiratory illness: From conventional microbiological methods to molecular detection (multiplex). Pathol Biol (Paris) 63: 69-73. [Crossref]


