Tricetin is a type of flavonoid that plays an important role in anti-cancer activity. However, the protein targets and function mechanism of tricetin in HepG2 cells remain unclear, which greatly limits its clinical application. In this paper, tricetin was immobilized by 3D photo cross linking chip and the microfluidic environment was established. The SPR (surface plasmon resonance) technique was used to monitor the protein targets interacting with the tricetin on the surface of the chip. The target proteins captured by tricetin in HepG2 cell were identified by HPLC-MS (high performance liquid chromatography-mass pec trometry) method. Bioinformatics annotation and analysis of the obtained proteins showed that the VDR (vitamin D receptor) and PIM1 (Ser/Thr-protein kinase-1) were significantly enriched in the vitamin D receptor signalling pathway (Rich factor > 0.6), and thirty high affinity proteins mainly involved in pathways in Cancer, MAPK (mitogen-activated protein kinase) signalling pathway, TNF (tumour necrosis factor) signalling pathway, Osteoclast differentiation. Among them, four high score affinity target proteins, CYP1B1 (cytochrome P4501B1), VDR, PIM1 and GAA were screened by reverse molecular docking. Finally, the two target proteins, VDR and PIM1, which provided important theoretical support for tricetin in anti-liver cancer research were fully discussed.
Key words
Tricetin, HepG2 cells, SPR, protein targets, reverse molecular docking
Hepatocellular Carcinoma (HCC) is a common cancer that causes great mortality each year [1]. Chronic infection with hepatitis B virus is a major risk factor for disease in Southeast Asia and Africa, while chronic hepatitis C infection is a major risk factor for disease in Europe [2,3]. In recent years, with increased life expectancy, the number of elderly HCC patients have also raised. At present, treatment and control measures for HCC mainly include surgical control, transplantation, percutaneous ethanol injection, radiation therapy and chemotherapy [4]. However, HCC is highly resistant to chemotherapy and is still ineffective for patients with advanced disease.
Flavonoids are polyphenols produced by secondary metabolism in plants [5]. They are ubiquitous in fruits, vegetables, tea and wine, which are considered to have positive impacts on human health [6-8]. Tricetin (5,7,3',4',5'-pentahydroxyflavone) (For the molecular structure of tricetin see Figure 1A, for the 3D structure see Figure 1B) is a kind of polyhydroxy flavone that originally found in pollen of Myrtaceae and honey of Eucalyptus [9-12], performs high activity in anti-inflammatory and anti-cancer [13,14]. Studies have shown that tricetin can cause cell cycle arrest in human breast cancer MCF-7 cells in the G2/M phase and induce a series of apoptotic responses, suggesting that tricetin may be a promising anti-breast cancer drug [14]. Besides, studies have found that tricetin induces HepG2 cell death by triggering the mitochondria and DR5 (death receptor 5) apoptotic pathway to achieve the purpose of treating liver cancer [15]. Incubation of HepG2 cells with tricetin resulted in decreased glutathione and ROS (reactive oxygen species) production, suggesting that tricetin-induced hepatoma cell death may be triggered by ROS. Intrinsic and extrinsic apoptotic pathways induce cancer cell death, suggesting the potential of tricetin for treatment of liver cancer [15]. In the anti-hepatocarcinoma process of tricetin, the intracellular action mechanism of tricetin in HepG2 has been verified and unfolded around several traditional pathways and proteins (e.g. PI3K/Akt, Bcl-2, JNK), which greatly limited the research breakthrough on tricetin. The emergence of SPR provides a providential opportunity for the study of drug targets due to its label-free and real-time detection properties [16].
Figure 1. (A). Chemical structure of tricetin. (B). 3D structure of tricetin
Considering the fact that it can comprehensively provide the binding strength and specificity information of the available molecular drugs and the targets, SPR technology was used to capture protein targets of tricetin in HepG2 cells. In addition, reverse molecular docking, a computer-assisted drug design technique that automatically interfaces with protein databases to find potential protein targets for small molecule drugs, can accelerate the understanding of the functional mechanisms of natural compounds [17]. In this paper, the binding proteins targets were screened by SPR technology and identified by in situ mass spectrometry. Obtained target proteins were analyzed by bioinformatic tools, and reversed molecular docking was used to screen the most likely anti-hepatocarcinoma protein targets and molecular mechanisms of tricetin. Overall, this study lays important theoretical foundation for tricetin as potential anti-liver cancer candidates (Flow chart see Figure 2).
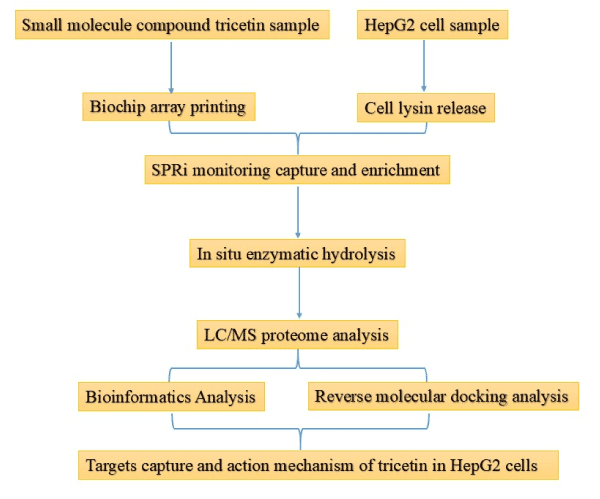
Figure 2. Flow chart of the study
Protein targets capture
After installing the chip on the SPR biochip analysis system, we adjust the test baseline and regenerate the surface of the chip for three times. The circulating regenerating liquid is Gly•HCl (pH 2.0), the flow rate is 3 μl/s, duration time is 300 s and the carrier buffer is 1 × PBST (0.05% Tween-20). The chip surface was blocked with 100 μg/ml BSA at a flow rate of 3 μl/s for 300 s. During the SPR test, the mobile phase was HepG2 cell lysate, and the surface stationary phase of the chip was tricetin. The SPR biochip analyser was used to monitor the binding of the immobilized molecules to the protein targets in the lysate (Figure 3). The operation time of 0 s ~ 260 s means pre-washing of the system and the surface of the chip was infiltrated in the buffer. The resonance intensity at this time is about 0 RU. The operation time of 260 s ~ 520 s means immobilized tricetin to capture protein targets in the lysate. And this region (260 s ~ 520 s) also indicates that the non-spotted region is also bound by a van der Waals force and hydrophobic interaction to bind certain proteins, but the signal in the non-spot area is significantly different from the spotted zone. In this area (520 s ~ 820 s), the chip was washed to remove non-specific adhesion proteins. After that, the molecular specific binding protein targets remain on the surface of the chip, while the non-binding molecules and non-specific molecules gradually leave, and the resonance intensity decreases to reach the plateau (~ 613.75 RU). With the non-specific binding targets were gradually cleaned, the resonance intensity of the background value gradually fell back to the baseline level (~ 28.36 RU), and the background noise of the chip returned to normal.
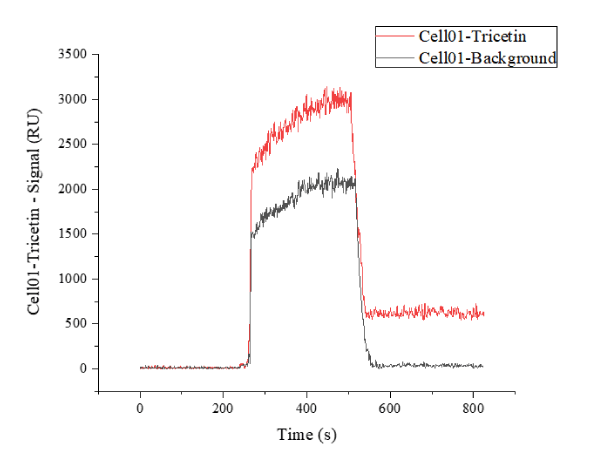
Figure 3. Tricetin spotting area signal curve (red) indicates the signal change of the compound spotting area on the chip, and the background noise signal curve (black) indicates the signal change of the unspotted area
HPLC-MS/MS identification of protein targets
After the test, the chip was subjected to in situ enzymatic hydrolysis in a monitoring device, and then the protein species enriched on the surface of the chip were identified by HPLC-MS. After analysis, it was confirmed that a total of 53 proteins were obtained (see Table 1 for detailed protein information, and Supplementary Table 1 for the original information of the proteins). Among them:
Table 1. Details of protein information enriched on the surface of the chip identified by HPLC-MS
Entry |
Protein name |
Abb |
Score |
Coverage |
Proteins |
Unique Peptides |
PSMs |
MW [kDa] |
P11309 |
Serine/threonine - protein kinase pim - 1 OS=Homo sapiens GN=PIM1 PE=1 SV=3 - [ PIM1_HUMAN ] |
PIM1 |
1953.94 |
62.92 |
1 |
6 |
82 |
35.6 |
Q9Y3R4 |
Sialidase - 2 OS=Homo sapiens GN=NEU2 PE=2 SV=3 - |
NEU2 |
1690.44 |
51.61 |
3 |
5 |
57 |
42.2 |
P28482 |
Mitogen - activated protein kinase 1 OS=Homo sapiens GN=MAPK1 PE=1 SV=1 - [ MK01_HUMAN ] |
MAPK1 |
1669.84 |
48.09 |
3 |
4 |
43 |
41.3 |
P22303 |
Acetylcholinesterase OS=Homo sapiens GN=ACHE PE=1 SV=1 - |
ACHE |
1582.34 |
51.8 |
2 |
8 |
93 |
67.7 |
O14920 |
Inhibitor of nuclear factor kappa - B kinase subunit beta OS=Homo sapiens GN=IKBKB PE=1 SV=3 - [ IKKB_HUMAN ] |
IKBKB |
1547.14 |
47.65 |
4 |
9 |
97 |
86.5 |
Q15628 |
Tumor necrosis factor receptor type 1 - associated DEATH domain protein OS=Homo sapiens GN=TRADD PE=1 SV=3 - [ TRADD_HUMAN ] |
TRADD |
1543.7 |
44.46 |
1 |
4 |
40 |
34.2 |
P04839 |
Cytochrome b - 245 heavy chain OS=Homo sapiens GN=CYBB PE=1 SV=1 - [ CY24B_HUMAN ] |
CYBB |
1397.24 |
40.52 |
4 |
6 |
56 |
65.3 |
O94782 |
Ubiquitin carboxyl - terminal hydrolase 1 OS=Homo sapiens GN=USP1 PE=1 SV=2 - [ UBP1_HUMAN ] |
USP1 |
1395.82 |
40.39 |
3 |
8 |
75 |
88.2 |
P17252 |
Protein kinase C alpha type OS=Homo sapiens GN=PRKCA PE=2 SV=1 - [ KPCA_HUMAN ] |
PRKCA |
1358.88 |
45.21 |
2 |
8 |
83 |
76.7 |
P10253 |
Lysosomal alpha - glucosidase OS=Homo sapiens GN=GAA PE=2 SV=2 - [ LYAG_HUMAN ] |
GAA |
1350.3 |
40.78 |
3 |
9 |
86 |
105.3 |
P68871 |
Hemoglobin subunit beta OS=Homo sapiens GN=HBB PE=1 SV=3 - |
HBB |
1345.78 |
38.22 |
1 |
2 |
17 |
15.9 |
P11473 |
Vitamin D3 receptor OS=Homo sapiens GN=VDR PE=2 SV=3 - |
VDR |
1306.45 |
39.63 |
2 |
5 |
46 |
48.2 |
P00352 |
Retinal dehydrogenase 1 OS=Homo sapiens GN=ALDH1A1 PE=1 SV=3 - [ AL1A1_HUMAN ] |
ALDH1A1 |
1301.25 |
35.48 |
1 |
5 |
42 |
54.8 |
P08047 |
Transcription factor Sp1 OS=Homo sapiens GN=SP1 PE=1 SV=3 - |
SP1 |
1264.92 |
39.72 |
1 |
8 |
74 |
80.6 |
P21964 |
Catechol O - methyltransferase OS=Homo sapiens GN=COMT PE=1 SV=3 - |
COMT |
1171.2 |
35.76 |
1 |
3 |
25 |
30 |
P26038 |
Moesin OS=Homo sapiens GN=MSN PE=1 SV=1 - [ MOES_HUMAN ] |
MSN |
1164.79 |
37.2 |
1 |
5 |
44 |
67.8 |
P51570 |
Galactokinase OS=Homo sapiens GN=GALK1 PE=2 SV=3 - [ GALK1_HUMAN ] |
GALK1 |
1143.63 |
37.43 |
3 |
4 |
35 |
42.2 |
P08684 |
Cytochrome P450 3A4 OS=Homo sapiens GN=CYP3A4 PE=1 SV=2 - [ CP3A4_HUMAN ] |
CYP3A4 |
1137.44 |
33.06 |
3 |
4 |
31 |
57.3 |
P11233 |
Ras - related protein Ral - A OS=Homo sapiens GN=RALA PE=1 SV=3 - [ RALA_HUMAN ] |
RALA |
1122.25 |
34.49 |
4 |
2 |
15 |
23.5 |
P11474 |
Steroid hormone receptor ERR1 OS=Homo sapiens GN=ESRRA PE=2 SV=1 - [ ERR1_HUMAN ] |
ESRRA |
1102.32 |
36.23 |
2 |
4 |
34 |
45.5 |
B2RXH2 |
Lysine - specific demethylase 4E OS=Homo sapiens GN=KDM4E PE=2 SV=1 - [ KDM4E_HUMAN ] |
KDM4E |
1089.8 |
29.64 |
3 |
4 |
29 |
56.8 |
P10636 |
Microtubule - associated protein tau OS=Homo sapiens GN=MAPT PE=1 SV=1 - [ TAU_HUMAN ] |
MAPT |
1077.12 |
34.61 |
3 |
7 |
58 |
78.9 |
Q13332 |
Receptor - type tyrosine - protein phosphatase S OS=Homo sapiens GN=PTPRS PE=1 SV=1 - [ PTPRS_HUMAN ] |
PTPRS |
1075.78 |
30.19 |
2 |
14 |
106 |
217 |
Q6NUS8 |
UDP - glucuronosyltransferase 3A1 OS=Homo sapiens GN=UGT3A1 PE=2 SV=3 - [ UD3A1_HUMAN ] |
UGT3A1 |
1066.3 |
35.19 |
2 |
5 |
42 |
59.1 |
Q16678 |
Cytochrome P450 1B1 OS=Homo sapiens GN=CYP1B1 PE=1 SV=3 - [ CP1B1_HUMAN ] |
CYP1B1 |
1063.93 |
28.87 |
1 |
4 |
28 |
60.8 |
P16050 |
Arachidonate 15 - lipoxygenase OS=Homo sapiens GN=ALOX15 PE=1 SV=2 - [ LOX15_HUMAN ] |
ALOX15 |
1060.22 |
32.16 |
1 |
6 |
47 |
74.8 |
P25963 |
NF - kappa - B inhibitor alpha OS=Homo sapiens GN=NFKBIA PE=1 SV=1 - [ IKBA_HUMAN ] |
NFKBIA |
1042.91 |
31.91 |
2 |
3 |
23 |
35.6 |
Q9NPD5 |
Solute carrier organic anion transporter family member 1B3 OS=Homo sapiens GN=SLCO1B3 PE=2 SV=3 - [ SO1B3_HUMAN ] |
SLCO1B3 |
1034.65 |
28.35 |
1 |
5 |
35 |
77.4 |
O43318 |
Mitogen - activated protein kinase kinase kinase 7 OS=Homo sapiens GN=MAP3K7 PE=1 SV=1 - [ M3K7_HUMAN ] |
MAP3K7 |
1015.45 |
33.58 |
3 |
6 |
48 |
67.1 |
P08631 |
Tyrosine - protein kinase HCK OS=Homo sapiens GN=HCK PE=1 SV=1 - [ HCK_HUMAN ] |
HCK |
1004.34 |
32.94 |
3 |
4 |
31 |
59.6 |
P10589 |
COUP transcription factor 1 OS=Homo sapiens GN=NR2F1 PE=1 SV=2 - [ COT1_HUMAN ] |
NR2F1 |
993.01 |
29.39 |
3 |
3 |
21 |
46.1 |
Q9UBT6 |
DNA polymerase kappa OS=Homo sapiens GN=POLK PE=2 SV=3 - [ POLK_HUMAN ] |
POLK |
940.17 |
25.2 |
1 |
5 |
32 |
98.8 |
Q9Y6L6 |
Solute carrier organic anion transporter family member 1B1 OS=Homo sapiens GN=SLCO1B1 PE=2 SV=1 - [ SO1B1_HUMAN ] |
SLCO1B1 |
933.99 |
24.97 |
4 |
4 |
25 |
76.4 |
P03372 |
Estrogen receptor OS=Homo sapiens GN=ESR1 PE=1 SV=3 - [ ESR1_HUMAN ] |
ESR1 |
924.63 |
29.83 |
4 |
5 |
37 |
66.2 |
Q9UNQ0 |
ATP - binding cassette sub - family G member 2 OS=Homo sapiens GN=ABCG2 PE=1 SV=2 - [ ABCG2_HUMAN ] |
ABCG2 |
866.1 |
25.75 |
1 |
5 |
33 |
72.3 |
Q00534 |
Cyclin - dependent kinase 6 OS=Homo sapiens GN=CDK6 PE=1 SV=1 - [ CDK6_HUMAN ] |
CDK6 |
848 |
24.65 |
3 |
2 |
12 |
36.9 |
Q99714 |
3 - hydroxyacyl - CoA dehydrogenase type - 2 OS=Homo sapiens GN=HSD17B10 PE=1 SV=1 - [ HCD2_HUMAN ] |
HSD17B10 |
827.57 |
23.94 |
3 |
2 |
12 |
26.9 |
P06576 |
ATP synthase subunit beta, mitochondrial OS=Homo sapiens GN=ATP5F1B PE=1 SV=3 - [ ATPB_HUMAN ] |
ATP5F1B |
783.23 |
23.08 |
2 |
3 |
18 |
56.5 |
P48736 |
Phosphatidylinositol 4,5 - bisphosphate 3 - kinase catalytic subunit gamma isoform OS=Homo sapiens GN=PIK3CG PE=1 SV=2 - [ PK3CG_HUMAN ] |
PIK3CG |
724.3 |
21.29 |
1 |
6 |
35 |
126.4 |
O00303 |
Eukaryotic translation initiation factor 3 subunit F OS=Homo sapiens GN=EIF3F PE=1 SV=3 - [ EIF3F_HUMAN ] |
EIF3F |
566.97 |
15.84 |
1 |
2 |
8 |
37.5 |
P16083 |
Ribosyldihydronicotinamide dehydrogenase [quinone] OS=Homo sapiens GN=NQO2 PE=2 SV=3 - [ NQO2_HUMAN ] |
NQO2 |
524.91 |
15.96 |
3 |
1 |
4 |
25.9 |
P68400 |
Casein kinase II subunit alpha OS=Homo sapiens GN=CSNK2A1 PE=1 SV=1 - [ CSK21_HUMAN ] |
CSNK2A1 |
522.88 |
14.19 |
3 |
2 |
8 |
45.1 |
P35869 |
Aryl hydrocarbon receptor OS=Homo sapiens GN=AHR PE=1 SV=1 - [ AHR_HUMAN ] |
AHR |
517.17 |
16 |
2 |
4 |
19 |
96.1 |
Q8N1C3 |
Gamma - aminobutyric acid receptor subunit gamma - 1 OS=Homo sapiens GN=GABRG1 PE=1 SV=2 - [ GBRG1_HUMAN ] |
GABRG1 |
268.52 |
8.74 |
2 |
1 |
2 |
53.5 |
P10632 |
Cytochrome P450 2C8 OS=Homo sapiens GN=CYP2C8 PE=1 SV=2 - [ CP2C8_HUMAN ] |
CYP2C8 |
197.75 |
5.7 |
3 |
1 |
2 |
55.8 |
P16152 |
Carbonyl reductase [NADPH] 1 OS=Homo sapiens GN=CBR1 PE=1 SV=2 - [ CBR1_HUMAN ] |
CBR1 |
154.04 |
4.51 |
3 |
1 |
1 |
30.3 |
P05177 |
Cytochrome P450 1A2 OS=Homo sapiens GN=CYP1A2 PE=2 SV=1 - [ CP1A2_HUMAN ] |
CYP1A2 |
150.03 |
4.57 |
3 |
1 |
1 |
58.4 |
O15244 |
Solute carrier family 22 member 2 OS=Homo sapiens GN=SLC22A2 PE=1 SV=1 - [ S22A2_HUMAN ] |
SLC22A2 |
133.61 |
4.26 |
3 |
1 |
1 |
62.5 |
O15245 |
Solute carrier family 22 member 1 OS=Homo sapiens GN=SLC22A1 PE=2 SV=3 - [ S22A1_HUMAN ] |
SLC22A1 |
126.01 |
4.01 |
4 |
1 |
1 |
61.1 |
O75751 |
Solute carrier family 22 member 3 OS=Homo sapiens GN=SLC22A3 PE=2 SV=2 - [ S22A3_HUMAN ] |
SLC22A3 |
79.71 |
2.58 |
2 |
1 |
1 |
61.2 |
P07900 |
Heat shock protein HSP 90 - alpha OS=Homo sapiens GN=HSP90AA1 PE=1 SV=1 - [ HS90A_HUMAN ] |
HSP90AA1 |
69.2 |
2.09 |
2 |
1 |
1 |
84.6 |
P60709 |
Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 - [ ACTB_HUMAN ] |
ACTB |
52.36 |
1.46 |
1 |
1 |
1 |
41.7 |
P02768 |
Serum albumin OS=Homo sapiens GN=ALB PE=2 SV=2 - [ ALBU_HUMAN ] |
ALB |
17.16 |
0.47 |
1 |
1 |
1 |
69.3 |
(1) Score > 1000 targets (30 in total, blue region): characterized by fast binding and high affinity, such target proteins are usually associated with acute efficacy, acute toxicity and drug metabolism.
(2) Targets with 200 < Score < 1000 (14 in total, purple region): characterized by moderate binding speed and affinity, such targets are usually associated with chronic efficacy.
(3) Targets with 100 < Score < 200 (5 in total, olive region): characterized by slow binding and low affinity, such targets are usually associated with drug delivery, plasma concentration maintenance, bypass depending on the efficacy of the drug, the corresponding target may not be the main pharmacodynamic target of the drug, but such target may be the subject of new drug use and new drug activity.
(4) Score < 100 is a non-specific binding (4 in total, dark gray area), and most of this protein is a carrier protein and a cytoskeletal protein, which can be excluded.
Bioinformatics analysis of protein targets
DAVID analysis tool was used to annotate the 30 high-affinity binding proteins, GO (Gene Ontology) functional annotation was shown in Figure 4. The results suggested that VDR and PIM1 were the most abundant in the VDR signalling pathway (Rich factor > 0.6), followed by MAP3K7 and IKBKB (IκB kinase beta), which were abundant in the IκB kinase complex (Rich factor = 0.2). The degree of enrichment is large. There are fourteen genes that are most abundantly enriched in cytosol (Q value is close to 0), which are PRKCA (protein kinase C alpha), HCK (haematopoietic cell kinase), NFKBIA (Nuclear factor kappa-B), COMT (Catechol-O-methyltransferase), TRADD (TNFR1-associated death domain protein), MAP3K7, ALDH1A1 (aldehyde dehydrogenase encoded by Aldh1a1), MAPK1, GALK1 (Galactokinase), ALOX15 (12/15-lipoxygenase gene), MAPT (microtubule-associated protein tau gene), IKBKB, NEU2 (neuraminidase 2) and HBB. Fourteen genes are significantly enriched in the plasma membrane, namely PRKCA, ACHE, PIM1, PTPRS (Protein tyrosine phosphatase σ), NFKBIA, COMT, TRADD, MAP3K7, SLCO1B3, CYBB, ALOX15, MAPT, RALA and MSN respectively.
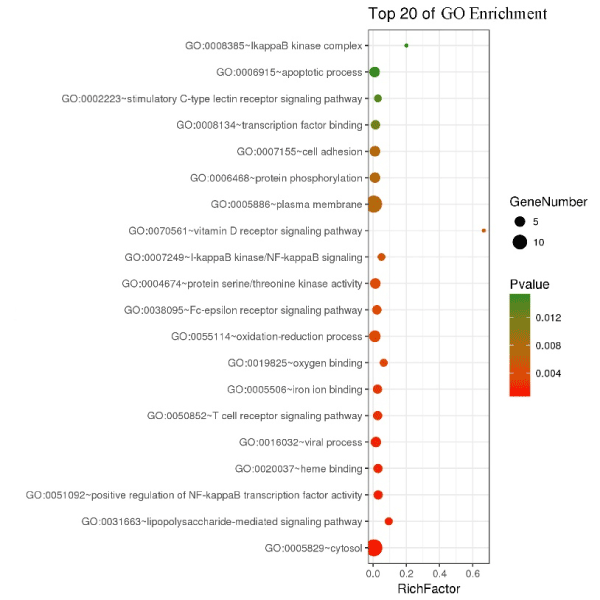
Figure 4. GO enrichment 30 high affinity protein targets of tricetin in HepG2 cell. The y coordinate indicates the function names of the GO analysis, and the horizontal axis indicates the Rich factor (the ratio of the number of differentially expressed genes in the GO entry to the number of total annotated genes in the GO entry. Larger Rich factor value indicates higher degree of enrichment). The size of the dot indicates the number of differentially expressed genes, and the color of the dot corresponds to the P value, which is the P value after multiple hypothesis test and correction and has the range of [0, 1]. A lower P value indicates that the enrichment is more significant
The KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment was shown in Figure 5. The results indicate that the 30 high affinity binding proteins of HepG2 cells were mainly involved in the pathways in Cancer, MAPK signalling pathway, TNF signalling pathway and Osteoclast differentiation. The five proteins in pathways in cancer are PRKCA, MAPK1, NFKBIA, RALA and IKBKB; five proteins of MAPK signalling pathway, namely MAP3K7, PRKCA, MAPK1, MAPT and IKBKB; five proteins of TNF signalling pathways are MAP3K7, MAPK1, NFKBIA, IKBKB and TRADD; the five proteins of Osteolast differentiation are MAP3K7, MAPK1, CYBB, NFKBIA and IKBKB.
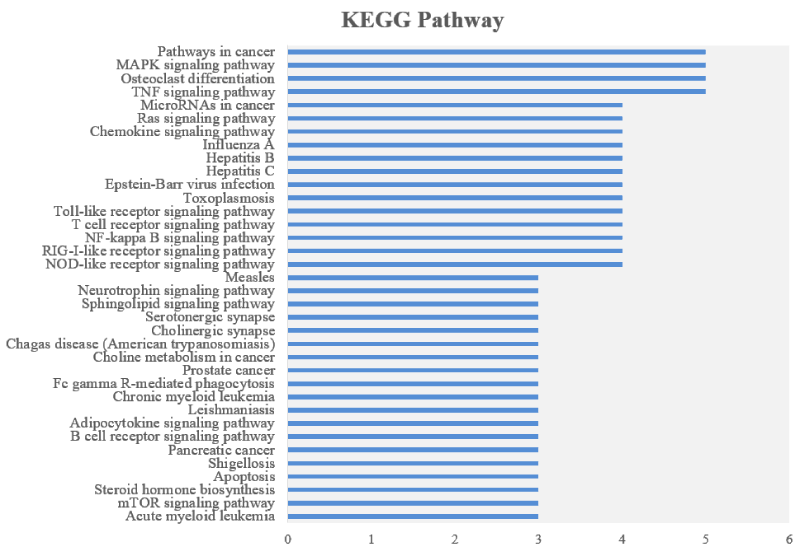
Figure 5. KEGG analysis of 30 high affinity protein targets of tricetin in HepG2 cell. The y axis represents the KEGG pathway entry and the x axis indicates the protein number of the entry
Reverse molecular docking
In order to quickly and accurately screen out the target proteins of tricetin in HepG2 cells, and to speed up the molecular mechanism research of tricetin, reverse molecular docking experiments on 14 high-affinity binding proteins published in the PBD database were performed. And the results were shown in Figure 6. Auto Grid was used to execute lattice calculation on the active site of the proteins and all values are default. Docking results were arranged according to the affinity energy, which reflects the affinity between the ligand and the proteins. The lower the energy of the docking, the higher the affinity force (Table 2). The protein targets with high affinity were screened four proteins,CYP1B1, VDR, PIM1, and GAA.
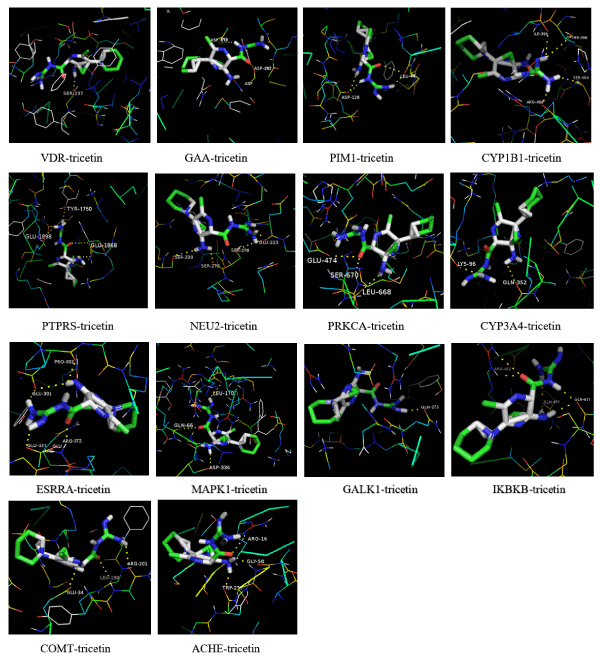
Figure 6. Reverse molecular docking experiments on 14 high-affinity binding proteins published in the PBD database
Table 2. Reverse molecular docking information summary
No. |
Protein name |
PDB ID |
Number of hydrogen bonds |
Interaction residues |
Affinity
(kcal/mol) |
1 |
VDR |
3W0Y |
1 |
SER-237 |
-10.7 |
2 |
GAA |
5KZW |
2 |
ASP-616;ASP-282 |
-9.6 |
3 |
PIM1 |
2O3P |
4 |
LEU-44; LEU-44;
ASP-128; ASP-128 |
-9.5 |
4 |
CYP1B1 |
6IQ5 |
4 |
ILE-399;THR-398
SER-464;ARG-468 |
-9.5 |
5 |
PTPRS |
2FH7 |
4 |
TYR-1750;GLU-1898;
GLU-1868; GLU-1868 |
-8.7 |
6 |
NEU2 |
1SNT |
4 |
GLU-223; SER-278;
SER-278; SER-339 |
-8.7 |
7 |
PRKCA |
3IW4 |
3 |
SER-670; GLU-474;
IEU-668 |
-8.6 |
8 |
CYP3A4 |
3NXU |
2 |
LYS-96; GLN-352 |
-8.6 |
9 |
ESRRA |
3K6P |
4 |
GLU-331; GLU-301
ARG-372; PRO-302 |
-8.4 |
10 |
MAPK1 |
1TVO |
2 |
GLN-66; LEU-170 |
-8.3 |
11 |
GALK1 |
6Q90 |
1 |
GLN-273 |
-8.0 |
12 |
IKBKB |
4KIK |
6 |
GLN-451; GLN-451;
GLN-611; GLN-611;
ARG-452; ARG-452 |
-7.3 |
13 |
COMT |
3A7E |
3 |
GLU-34; ARG-201;
LEU-198 |
-7.0 |
14 |
ACHE |
4PQE |
3 |
TRP-271; ARG-16;
GLY-58 |
-5.4 |
Materials and reagents
Hepatoma cell line HepG2 was purchased from Procell company. Tricetin was purchased from Extrasynthese (Genay, France), dissolved in DMSO (Sigma Aldrich, St. Louis, MO) and stored at -20℃.
Cell culture
The HepG2 cells were taken out from the liquid nitrogen tank, and quickly dissolved in a 37℃ constant temperature water bath, then transferred to a sterile centrifuge tube, centrifuged at 200 g for 5 minutes. Then the supernatant was discarded and the cells were resuspended and cultured in pre-warmed DMEM (Invitrogen, Carlsbad, CA) medium containing 1% penicillin and streptomycin, 10% FBS (GIBCO, Gaithersburg, MD) at 37℃containing 5% CO2 constant temperature incubator.
Liquid Chromatography Mobile Phase A: 5% ACN, 0.1% aqueous formic acid, pH = 2.5 (chromatographically pure formic acid); liquid chromatography mobile phase B: 90% ACN, 0.1% aqueous formic acid, pH = 2.5 (Chromatographic pure formic acid. The ion source spray voltage was 2.0 kV, the mass spectrometer heating capillary was set to 250℃, and the data-dependent mode was used to automatically switch between MS and MS/MS. The full scan MS Orbitrap was used for scanning with 90 min at scan range of m/z 350-1600, and a resolution of 70,000 (m/z 200). The parent ion is screened using a quadrupole, and then the parent ion of the cascade (MS/MS) fragmentation condition is fragmented using HCD (High Energy C-trap dissociation) and scanned with Orbitrap. And the scan resolution is 17,500; the scan range is automatically controlled according to the parent ion mass-to-charge ratio. MS/MS scans were performed on the top 15 ions of intensity. The parent ion selection window is set to 2Da. The MS/MS acquisition was not performed for ions with a single charge and an unknown charge number, and the dynamic exclusion was set to one MS/MS per parent ion for 30s. MS/MS uses high purity N2 with 27% collision energy. MS data was collected by XcaliburTM Software (Thermo Scientific, version 2.4.5).
MS data was searched by Mascot algorithm using Proteome Discoverer (Thermo Fisher Scientific, version 1.7) analysis software. The search database was UniProtKB/Swiss-Prot protein database. In order to reduce the false positive result, a database (Decoy database) containing all protein inversion sequences was added. The searched species were: Homo Sapiens (Human), database version: 2018_07_02, database capacity (number of protein records): 161,567 records. Search parameters were set as follows: Trypsin, full enzyme digestion, maximum missed cut is 2. The variable modification is the methionine (M) oxidation and deamidation (NQ) of the peptide. Single isotope mode was set, peptide mass error is 10 ppm, and fragment ion mass error is 0.05 Da. Peptide results used the Percolator algorithm to control peptide false positive rates (FDR) below 1%.
DAVID (https://david.ncifcrf.gov/) was used to annotate protein (GO) and KEGG analysis. OmicShare (http://www.omicshare.com/tools/Home/Soft/senior) online tools were used to draw the GO enriched bubble map.
First, ChemDraw was used to draw the 3D structure of tricetin. The crystal structures of high affinity protein targets were downloaded from the RCSB Protein Data bank (PDB) (http://www.rcsb.org/). All the heteroatoms, the water of crystallization, and the ligands carried in the crystal structure were all removed, and polar hydrogen is added. All of the above optimization processes were done on AutoDock Tools-1.5.6 and PyMOL 1.5.0.4 software. Finally, use the command prompt and PyMOL to analyse and plot the experimental results.
In the study of the interaction between small molecules and proteins, the kinetics of drug binding to the target has a lot to do with the effect of drug therapy. One of the main tasks of lead compound optimization is to increase the affinity of the drug candidate to the targets. It is necessary to fully understand the properties of candidate compounds and detailed kinetic parameters, which are important criteria for drug research and evaluation [18]. SPR biosensing technology has become an important tool in the field of drug research because of its high sensitivity, high throughput and label-free methodology. In this paper, 53 protein targets of tricetin in HepG2 cells were captured by SPR technology, and the high affinity 30 proteins was analyzed by bioinformatics. These proteins were mainly involved in vitamin D receptor signalling pathway and pathway in Cancer. CYP1B1, VDR, PIM1 and GAA were screened by reverse molecular docking, according to literature mining, VDR and PIM proteins may be the main potential targets of tricetin action in HepG2.
The vitamin D receptor (VDR) is a member of the nuclear receptor family of transcription factors [19]. In humans, the vitamin D receptor is encoded by the vdr gene. The downstream target of this nuclear hormone receptor is primarily involved in mineral metabolism, although the receptor regulates a variety of other metabolic pathways, such as those involved in immune responses and cancer [20]. Genetic analysis of VDR in HCC specimens of various etiology revealed a significant correlation between VDR polymorphism and alcohol-related HCC [21]. Furthermore, VDR expression in HCC tissues is increased compared to normal non-cancerous livers. In human hepatoma cell lines, activation of VDR by vitamin D or pharmacological ligands reduces cell proliferation [22,23]. In our study, the high-affinity proteins of the HepG2 cells that tricetin acts on mainly involved the VDR signalling pathway (Figure 3), and the reverse molecular docking results also showed that compared to other captured protein targets, the affinity between tricetin and VDR is the strongest (Figure 5). This series of evidence underlines the mechanism of tricetin's anti-liver cancer functional action to VDR.
PIM is a proto-oncogene belonging to the serine-threonine-protein kinase family [24]. Pim-1 is directly involved in the regulation of cell cycle progression and apoptosis [25], and is implicated in various cancers such as prostate cancer [26] and Burkitt's lymphoma [27]. Overexpression of PIM1 was observed in 39% of cases by immunohistochemical analysis of 56 human primary HCC samples. Under hypoxic conditions (1% O2), PIM1 was significantly up-regulated in multiple HCC cell lines compared to normoxia (20% O2). Compared with non-target controls, PIM1 silencing inhibited HCC cell invasion in vitro, decreased HCC cell proliferation in vitro, and reduced tumour growth and metastatic potential in vivo [28]. Knockdown of PIM1 significantly reduced glucose uptake by HCC cells and was associated with decreased levels of p-AKT and key molecules in the glycolytic pathway [28]. PIM kinase inhibitors are also considered to be a possible treatment for Alzheimer's disease [29]. Studies have shown that low molecular weight phenanthrene derivatives can promote cell cycle arrest by inhibiting Pim-3 activity [30]. Screening and design of Pim inhibitors can provide potential drug candidates for cancer therapy. In our study, reverse molecular docking results indicated that tricetin has a high affinity with Pim, suggesting the potential of tricetin to bind to Pim.
Cells lysis treatment
The collected HepG2 cell samples were centrifuged to concentrate the sample on the bottom of the EP tube, and 120 μl of PBS and a final concentration (v:v) of 1% Cocktail protease inhibitor (Thermo Fisher) were added, and the suspension was fully shaken. BWLS-17 lysate (BetterWays Inc.) was added at the ratio of 3:4 (v:v), and the cell sample was treated according to the "Syringe Jet Pyrolysis" SOP(Standard Operation Procedure) [31]. The treated sample was centrifuged at 16,000 g for 10 min at 4℃. The supernatant was taken for concentration determination (Thermo Fisher BCA Protein Assay Kit); the sample was adjusted for concentration using a 1x stock solution of lysate at a final concentration of 200 μg/ml.
Photo cross linking sensor chip production
A photo-crosslinking sensor chip (BetterWays Inc.) was taken out and returned to room temperature for 30 min. Tricetin was formulated in DMSO to a 100 mM solution. The compound solution was spotted on a designated area on the 3D photo cross linking sensor chip by a high throughput array printer. The printer set point spacing is 280 μm, the dot diameter is 180 μm, the chip surface contains 50 × 50 dot matrix (2,500 dots), the single point solution volume is 2.5 nl, and the spotting is repeated 3 times. The chip surface spotting amount is 18.75 ml (1.875 μM). During the period, it is strictly protected from light in N2 environment, and pressure is 1.05 ATMs. The printed chip is naturally dried in a low temperature dehumidification in the chip printer.
After drying, the chip was transferred to an ultraviolet light crosslinker (Amersham Life Science) for photo cross linking reaction at a wavelength of 365 nm, N2 ambient pressure of 1.20 ATMs. And irradiation procedure and parameters were set as follows: energy 9000 μW/cm, 2 min; pause 2 Min; energy 9000 μW/cm, 2 min; pause for 2 min; energy 2500 μW/cm, 15 min.
Protein targets capture
After installing the chip on the SPR biochip analysis system, we adjust the test baseline and regenerate the surface of the chip for three times. The circulating regenerating liquid is Gly•HCl (pH2.0), the flow rate is 3 μl/s, duration time is 300 s and the carrier buffer is 1×PBST (0.05% Tween-20). The chip surface was blocked with 100 μg/ml BSA at a flow rate of 3 μl/s for 300s. During the SPR test, the mobile phase was HepG2 cell lysate, and the surface stationary phase of the chip was tricetin.
In situ trypsin digestion
The chip protein was then subjected to in situ trypsin digestion. A final concentration of 10 mM DTT (DL-Dithiothreitol) solution to the chip was added and then reacting at 56℃ for 1 h. After reduction, a final concentration of 55 mM iodoacetamide solution was added. 30 μl 0.25 M tetraethylammonium bromide was used to wash for 2 ~ 3 times. Trypsin (trypsin stored in 50 mM acetic acid, storage concentration is 1 μg/μl) was added and incubated at 37℃ for 12 h. The digested peptide fraction was collected into a 1.5 ml tube, and then 30 μl of 0.5 M iodoacetamide was added.
In summary, the potential application of the natural product tricetin in the adjuvant treatment of angiogenesis, proliferation, progression and metastasis of malignant cells is receiving increasing attention. Defining the material basis of Chinese medicine is an important goal of the modernization of Chinese medicine. Due to the complex composition of traditional Chinese medicine, it is difficult to separate and purify the active ingredients. The traditional separation-identification-pharmacological test research method is expensive and inefficient. In particular, it is difficult for some pharmaceutical ingredients to be enriched in the amount required for pharmacological testing, and thus it is impossible to carry out related pharmacological experiments. In this paper, the binding proteins of tricetin to specific HepG2 cells was explored. The reverse molecular docking method was used to predict potential targets. Only the structure of the compound was used, and the activity and possible mechanism of the drug could be predicted based on its compatibility with known protein targets. According to the predicted results, targeted pharmacological experiments can be carried out to avoid blind attempts, and promoting the research process of modernization of natural products.
This work was supported by National Natural Science Foundation of China (31702186, 31872425, 31861143051) and the Natural Science Foundation of Jiangsu Province, China (BK20160509).
The authors declare that they have no conflict of interest.
View Supplementary Data
- McGlynn KA, London WT (2011) The global epidemiology of hepatocellular carcinoma: present and future. Clinics in liver disease 15: 223-243. [Crossref]
- Llovet JM, Burroughs A, Bruix J (2003) Hepatocellular carcinoma. Lancet 362: 1907-1917. [Crossref]
- El-Serag HB, Mason AC (1999) Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med 340: 745-750. [Crossref]
- Yokoyama I, Todo S, Iwatsuki S, Starzl TE (1990) Liver transplantation in the treatment of primary liver cancer. Hepatogastroenterology 37: 188-193. [Crossref]
- Lee KH (1999) Anticancer drug design based on plant-derived natural products. Journal of biomedical science 6: 236-250. [Crossref]
- Graf BA, Milbury PE, Blumberg JB (2005) Flavonols, flavones, flavanones and human health: epidemiological evidence. Journal of medicinal food 8: 281-290. [Crossref]
- Kocic B, Kitic D, Brankovic S (2013) Dietary flavonoid intake and colorectal cancer risk: evidence from human population studies. Journal of BUON 18: 34-43. [Crossref]
- Duthie GG, Duthie SJ, Kyle JA (2000) Plant polyphenols in cancer and heart disease: implications as nutritional antioxidants. Nutr Res Rev 13: 79-106. [Crossref]
- Yao L, Jiang Y, D'Arcy B, Singanusong R, Datta N, et al. (2004) Quantitative high-performance liquid chromatography analyses of flavonoids in Australian Eucalyptus honeys. Journal of agricultural and food chemistry 52: 210-214. [Crossref]
- Campos MG, Webby RF, Markham KR (2002) The unique occurrence of the flavone aglycone tricetin in Myrtaceae pollen. Z Naturforsch C 57: 944-946. [Crossref]
- Martos I, Ferreres F, Yao L, D'Arcy B, Caffin N, et al. (2000) Flavonoids in monospecific eucalyptus honeys from Australia. J Agric Food Chem 48: 4744-4748. [Crossref]
- Martos I, Ferreres F, Tomas-Barberan FA (2000) Identification of flavonoid markers for the botanical origin of Eucalyptus honey. J Agric Food Chem 48: 1498-1502. [Crossref]
- Geraets L, Moonen HJ, Brauers K, Wouters EF, Bast A, et al. (2007) Dietary flavones and flavonoles are inhibitors of poly (ADP-ribose) polymerase-1 in pulmonary epithelial cells. J Nutr 137: 2190-2195. [Crossref]
- Hsu YL, Uen YH, Chen Y, Liang HL, Kuo PL (2009) Tricetin, a dietary flavonoid, inhibits proliferation of human breast adenocarcinoma mcf-7 cells by blocking cell cycle progression and inducing apoptosis. J Agric Food Chem 57: 8688-8695. [Crossref]
- Hsu YL, Hou MF, Tsai EM, Kuo PL (2010) Tricetin, a dietary flavonoid, induces apoptosis through the reactive oxygen species/c-Jun NH2-terminal kinase pathway in human liver cancer cells. J Agric Food Chem 58: 12547-12556. [Crossref]
- Wang W, Fang Q, Hu Z (2016) High-Throughput peptide screening on a bimodal imprinting chip through MS-SPRi integration. Methods Mol Biol 1352: 111-125. [Crossref]
- Lee A, Lee K, Kim D (2016) Using reverse docking for target identification and its applications for drug discovery. Expert Opin Drug Discov 11: 707-715. [Crossref]
- Weber PC, Salemme FR (2003) Applications of calorimetric methods to drug discovery and the study of protein interactions. Curr Opin Struct Biol 13: 115-121. [Crossref]
- Moore DD, Kato S, Xie W, Mangelsdorf DJ, Schmidt DR, et al. (2006) International union of pharmacology. LXII. The NR1H and NR1I receptors: constitutive androstane receptor, pregnene X receptor, farnesoid X receptor alpha, farnesoid X receptor beta, liver X receptor alpha, liver X receptor beta, and vitamin D receptor. Pharmacol Rev 58: 742-759. [Crossref]
- Adorini L, Daniel KC, Penna G (2006) Vitamin D receptor agonists, cancer and the immune system: an intricate relationship. Curr Top Med Chem 6: 1297-1301. [Crossref]
- Falleti E, Bitetto D, Fabris C, Cussigh A, Fontanini E, et al. (2010) Vitamin D receptor gene polymorphisms and hepatocellular carcinoma in alcoholic cirrhosis. World J Gastroenterol 16: 3016-3024. [Crossref]
- Pourgholami MH, Akhter J, Lu Y, Morris DL (2000) In vitro and in vivo inhibition of liver cancer cells by 1,25-dihydroxyvitamin D3. Cancer Lett 151: 97-102. [Crossref]
- Ghous Z, Akhter J, Pourgholami MH, Morris DL (2008) Inhibition of hepatocellular cancer by EB1089: in vitro and in vive study. Anticancer Res 28: 3757-3761. [Crossref]
- Qian KC, Wang L, Hickey ER, Studts J, Barringer K, et al. (2005) Structural basis of constitutive activity and a unique nucleotide binding mode of human Pim-1 kinase. J Biol Chem 280: 6130-6137. [Crossref]
- Quan J, Zhou L, Qu J (2015) Knockdown of Pim-3 suppresses the tumorigenicity of glioblastoma by regulating cell cycle and apoptosis. Cell Mol Biol (Noisy-le-grand) 61: 42-50. [Crossref]
- Valdman A, Fang X, Pang ST, Ekman P, Egevad L (2004) Pim-1 expression in prostatic intraepithelial neoplasia and human prostate cancer. Prostate 60: 367-371. [Crossref]
- Ionov Y, Le X, Tunquist BJ, Sweetenham J, Sachs T, et al. (2003) Pim-1 protein kinase is nuclear in Burkitt's lymphoma: nuclear localization is necessary for its biologic effects. Anticancer res 23: 167-178. [Crossref]
- Leung CO, Wong CC, Fan DN, Kai AK, Tung EK, et al. (2015) PIM1 regulates glycolysis and promotes tumor progression in hepatocellular carcinoma. Oncotarget 6: 10880-10892. [Crossref]
- Velazquez R, Shaw DM, Caccamo A, Oddo S (2016) Pim1 inhibition as a novel therapeutic strategy for Alzheimer's disease. Mol Neurodegener 11: 52. [Crossref]
- Wang YY, Taniguchi T, Baba T, Li YY, Ishibashi H, et al. (2012) Identification of a phenanthrene derivative as a potent anticancer drug with Pim kinase inhibitory activity. Cancer sci 103: 107-115. [Crossref]
- Tummala RR (2004) SOP: what is it and why? A new microsystem-integration technology paradigm-Moore's law for system integration of miniaturized convergent systems of the next decade. IEEE Transactions on Advanced Packaging 27: 241-249.






