Abstract
We established new LC-MS/MS methodology of the target proteome of unknown proteins in cytoplasm of hippocampus for Drug Discovery of Alzheimer's Treatment. Quinone oxidoreductase2 (QR2) of the 25kDa protein and Pyridoxal kinase (PK) of the 37kDa protein are the target proteins of Alzheimer’s Disease. FK960 binding to QR2 and PK inhibits respectively over-expression of QR2 disturbing memory formation at the cortex and over-phosphorylation of the microtubule-associated Tau protein accumulated at neurofibrillary tangles in the brains of Alzheimer’s disease patients.
Key words
Alzheimer's treatment, pyridoxal kinase, quinone oxidoreductase2, FK960, ESI Ion Trap MS
Introduction
FK960, [N-(4-acetyl-1-piperazinyl)-p-fluorobenzamide monohydrate], a novel antidementia piperazine derivative, has been shown to reverse scopolamine-induced cognitive deficits in rats in vivo [1], to increase the magnitude of long-term potentiation in guinea pig hippocampus in vitro [2], and improve visual recognition memory in primates [3]. FK960 (100 nM) significantly increased the amplitude of the Excitatory postsynaptic potentials [4]. These studies implicated somatostatinergic, cholinergic, and serotonergic systems. We did not know what kinds of proteins FK960 bind to and act pharmacological effect. For new drug discovery, it is important how to search a small molecule such as FK960 or the small molecule ligands of nuclear receptor (NR) binding promoter DNAs to control transcription of pharmaceutical active proteins such as NF-AT, NF-kB, HDAC, IL-1, TNF and peroxisome [5].
We established new LC-MS/MS methodology of the target proteome of unknown proteins in cytoplasm of hippocampus for Drug Discovery of Alzheimer's Treatment. Quinone oxidoreductase2 (NAD(P)H menadione oxido reductase) of the 25kDa protein and Pyridoxal kinase of the 37kDa protein are the target proteins of Alzheimer’s Disease.
Method
Synthesis of FK960-immobilized Sepharose 4B
FK960-immobilized Sepharose 4B was synthesized as shown in Figure 1.

Figure1. Synthesis of FK960-immobilized Sepharose 4B
Isolation of FK960 binding proteins in cytoplasm of hippocampus
FK960 binding proteins in cytoplasm of hippocampus was collected from 20 mail Sprague Dawley rats fasted using FK960-immobilized Sepharose 4B incubating with or without FK960 in buffer (50mM Tris-HCl (pH7.6), 20% glycerol, 0.3M NaCl) and then SDS page electrophoresis. FK960 binding proteins bind FK960 more than FK960-immobilized Sepharose 4B and so FK960 specific binding proteins bind with FK960-immobilized Sepharose 4B in the case of incubating without FK960, but in the case of incubating with FK960, FK960 specific binding proteins do not bind with FK960-immobilized Sepharose 4B as shown the methodology isolating FK960 binding proteins in Figure 2.
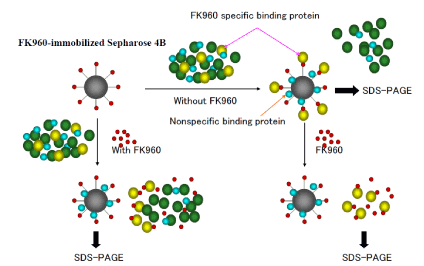
Figure 2. The methodology isolating FK960 binding proteins
LC-MS/MS analysis of FK960 binding proteins after in gel digestion of SDS page spots
FK960 binding protein bound with FK960-immobilized Sepharose 4B was eluted with FK960 after washing out another protein. The elute, the washout solution and the incubating solution with FK960 were compared the containing proteins using SDS page electrophoresis in both MES buffer and MOPS buffer in Figure 3 and Silver stain (in NuPAGE 4-12% Bis-Tris Gel MOPS buffer) and CBB stain in Figure 4. In the SDS page, the – marked bands contained 37kDa protein and 25kDa protein, but the + marked band did not contain them. 37kDa protein and 25kDa protein stained bands were cut off from the gel, and proteins were reduced by dithiothreitol, alkylated by iodoacetamide (SIGMA, St Louis, MO) and digested by Trypsin that had been modified for peptide sequencing (Roche, Mannheim, Germany). We carried out this in-gel digestion according to a previous report. (The total amount of precipitate was dissolved in 4mL of 6mol/L guanidine, 0.1mol/L tris and 1mmol/L EDTA buffer (pH8.3). After 300μmol DTT treatment to reduce S-S bond, 500μmol iodoacetic acid treatment to protect the produced SH group by carboxymethylation and a micro dialysis with a 6 kDa MW cut-off membrane, trypsin (2% w/w) was added to the solution and incubated at 37? for 4 hours and lyophilized.
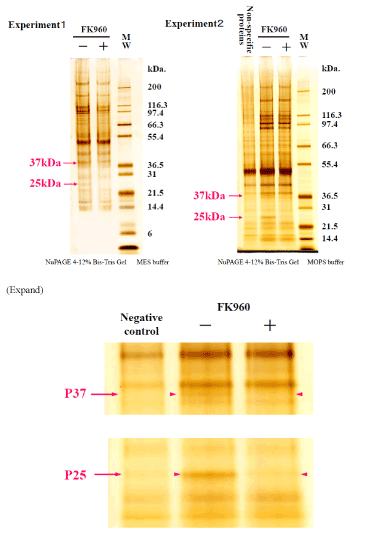
Figure 3. SDS page electrophoresis 1 isolating FK960 binding proteins + and -marks mean incubation with or without FK960
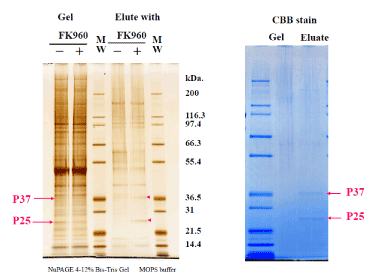
Figure 4. SDS page electrophoresis 2 isolating FK960 binding proteins + and -marks mean incubation with or without FK960
LC-MS/MS analysis of 37kDa protein and 25kDa protein were conducted after in gel digestion of the spot in SDS-page electrophoresis using nanoLC MAGIC 2002(Magic C18 column (0.75mm x150mm,3.5μm), Mobile phase ((A) 0.1%(v/v) AcOH in Water, (B) 0.1%(v/v) AcOH in acetonitrile, Gradient (0-5min (B;5%), 5-45min (B;5-45%), 45-55min (B;45-90%), 55-90min (B;90%),90-91min (B;90-5%),91-95 min (B;5%)), Flow rate (50 nL/min)) and ThermoFinnigan ion trap LCQ Deca MS under MS condition of analysis (Sheath gas (75), Auxiliary gas (15), Capillary temperature (300?), Scan mode (LC/ESI/MS/MS (positive mode, data dependent scan)).
Result
The high sensitive nanoLC-MS/MS measurement was conducted after in-gel digestion of both spots of 37kDa protein and 25kDa protein of SDS page electrophoresis. The following unknown 37kDa protein and 25kDa protein were identified by Bioworks Sequest (Thermo Electron) having high protein coverage, many repeat number of appearance, high quality of mass spectrum and high total score, and using rat and nr fasta data base. Mass Chromatogram (top), mass spectrum (middle) and MS/MS spectrum of 25kDa protein in SDS page electrophoresis are shown in Figure 5. Mass Chromatogram (top), mass spectrum (middle) and MS/MS spectrum of 37kDa protein in SDS page electrophoresis are shown in Figure 6.
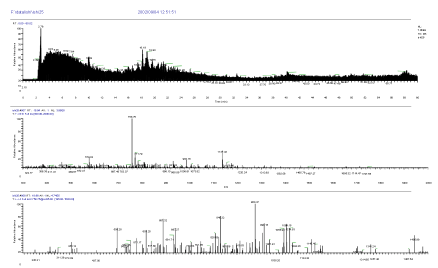
Figure 5. Mass Chromatogram (top), mass spectrum (middle) and MS/MS spectrum (bottom) of 25kDa protein in SDS page electrophoresis
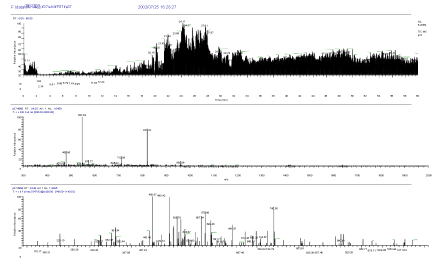
Figure 6. Mass Chromatogram (top), mass spectrum (middle) and MS/MS spectrum (bottom) of 37kDa protein in SDS page electrophoresis
The 25kDa protein was assigned Quinone oxidoreductase2 (NAD(P)H menadione oxidoreductase) that had never identified in rat and so used nr fasta database having protein coverage 52/231=22.5% by amino acid count, many repeat number of appearance, high total score in Figure 7 and high quality of MS/MS spectrum in Figure 8.
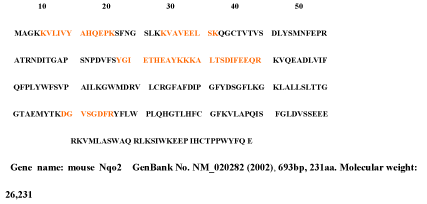
Figure 7. Quinone oxidoreductase2 of the 25kDa protein assigned by Bioworks Sequest soft using nr fasta database
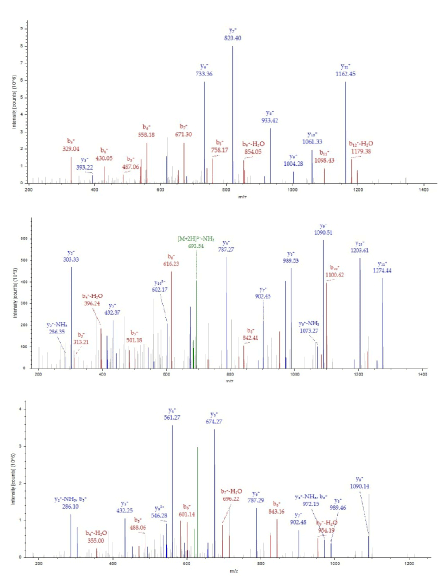
Figure 8. MS/MS spectrum of Quinone oxidoreductase2 of the 25kDa protein assigned by Bioworks Sequest soft using nr fasta database
The 37kDa protein was assigned Pyridoxal kinase having monoisotopic mass = 34867, Protein Coverage: 108/312 = 34.6% by amino acid count, 11857/34867 = 34.0% by mass, many repeat number of appearance, high total score in Figure 9 and high quality of MS/MS spectrum in Figure 10.

Figure 9. Pyridoxal kinase of the 37kDa protein assigned by Bioworks Sequest soft using nr fasta database
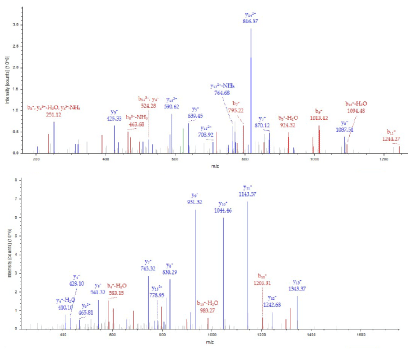
Figure 10. MS/MS spectrum of Pyridoxal kinase of the 37kDa protein assigned by Bioworks Sequest soft using nr fasta database
Discussion
We believe a new mechanism by FK960 binding 25kDa protein assigned Quinone oxido reductase2 (QR2) and FK960 binding 37kDa protein assigned Pyridoxal kinase (PK) to inhibit respectively over-expression of QR2 disturbing memory formation at the cortex [6] and over-phosphorylation of the microtubule-associated Tau [7,8] protein accumulated at neurofibrillary tangles in the brains of Alzheimer’s disease patients as described by many scientists [9-12].
We also believe this methodology is useful to find small molecule ligands of nuclear receptor (NR) binding promoter DNAs to control transcription of pharmaceutical active proteins such as NF-AT, NF-kB, HDAC, IL-1, TNF and peroxisome.
Acknowledgements
2021 Copyright OAT. All rights reserv
We would express our special thanks to Managing Editor Sandy Williams of Trends in Research, Dr. G.C. Stafford, Prof. D. Hunt, and Prof. R.G. Cooks for Inventors of ThermoFinnigan Ion Trap LCQ Deca MS who teach us MS good hand operation.
Conflicts of interest
The authors declare no conflict of interest.
References
- Matsuoka N, Satoh M (1998) FK960, a novel potential anti-dementia drug, augments long-term potentiation in mossy fiber-CA3 pathway of guinea-pig hippocampal slices. Brain Res 794: 248-254. [Crossref]
- Matsuoka N, Satoh M (1998) FK960, a novel potential anti-dementia drug, augments long-term potentiation in mossy fiber-CA3 pathway of guinea-pig hippocampal slices. Brain Res 794: 248-254. [Crossref]
- Matsuoka N and Aigner TG (1997) FK960, N-(4-acetyl-1-piperazinyl)-p- fluorobenzamide monohydrate], a novel potential anti-dementia drug, improves visual recognition memory in rhesus monkeys: comparison with physostigmine. J Pharmacol Exp Ther 280: 1201-1209.
- Hodgkiss JP, Kelly JS (2001) Effect of FK960, a putative cognitive enhancer, on synaptic transmission in CA1 neurons of rat hippocampus. J Pharmacol Exp Ther 297: 620-628. [Crossref]
- Tozuka Z, Kunisawa A (2018) Multi Omics by LC-MS/MS to search small molecule ligands of nuclear receptors to control transcription of pharmaceutical active proteins for drug discovery. Trends Res 1: 111.
- Akiva N, Rappaport AN, Jacob E, Sharma X, Inberg S, et al. (2015) Expression of Quinone Reductase-2 in the cortex is a muscarinic acetylcholine receptor-dependent memory consolidation constraint. J Neuroscience 35: 15568-15581.
- Aizawa H, Kawasaki H, Murofushi H, Kotani S, Suzuki K, et al. (1989) A common amino acid sequence in 190-kDa microtubule-associated protein and tau for the promotion of microtubule assembly. J Biol Chem 264: 5885-5890. [Crossref]
- Aizawa H, Kawasaki H, Murofushi H, Kotani S, Suzuki K, et al. (1989) A common amino acid sequence in 190-kDa microtubule-associated protein and tau for the promotion of microtubule assembly. J Biol Chem 264: 5885-5890. [Crossref]
- Aizawa H, Kawasaki H, Murofushi H, Kotani S, Suzuki K, et al. (1989) A common amino acid sequence in 190-kDa microtubule-associated protein and tau for the promotion of microtubule assembly. J Biol Chem 264: 5885-5890. [Crossref]
- Kimura T, Ishiguro K, Hisanaga S (2014) Physiological and pathological phosphorylation of tau by Cdk5. Front Mol Neurosci 7: 65. [Crossref]
- Crowther RA, Goedert M (2000) Abnormal tau-containing filaments in neurodegenerative diseases. J Struct Biol 130: 271-279. [Crossref]
- Lee VM, Kenyon TK, Trojanowski JQ (2005) Transgenic animal models of tauopathies. Biochim Biophys Acta 1739: 251-259. [Crossref]
- Otth C, Concha II, Arendt T, Stieler J, Schliebs R, et al. (2002) AbetaPP induces cdk5-dependent tau hyperphosphorylation in transgenic mice Tg2576. J Alzheimers Dis 4: 417-430. [Crossref]










