We have recently observed that one-year use of high fat diet (HFD) that induced mild ketonemia lead to better learning and memory, larger hippocampi volumes without any changes to cortical volumes, as well as higher concentrations of total NAA (tNAA: N-acetylaspartate and N-acetylaspartateglutame; marker of neuronal viability), total Cho (tCho: Glycerophosphocholine +Phosphocholine, which are believed to be primarily involved in cell membrane breakdown and synthesis) and total Cr (tCr: creatine + phospo-creatine – involved in cell bioenergetics). However, the spectroscopic results may have been driven by specific processing procedures used by LC Model, thus we needed to use a different software to assure the obtained results are independent of processing procedure. TARQUIN (Wilson et al, 2011) [1] is an open source alternative that was demonstrated to work comparably well to LCModelTM with wide range of 1.5T and 3.0T proton spectra. However, it has not been used to process proton, animal spectra acquired at 7.0T. Here, we 1) created basis sets for TARQUIN to work with spectra obtained at 7T, 2) reanalyzed the data, and finally 3) compared performance of TARQUIN and LCModelTM for single voxel hippocampal and anterior cingulate cortex spectra obtained from 50 one-month Wistar rats at 7T, and later, when they were one year old (n=47). Two different basis sets were proposed: one based on basis set used by LCModelTM, and the other one adapted from a basis set established for 16.1T. Given the intrinsic differences in processing between LCModelTM and TARQUIN, we evaluated quality of fit (Q) and performed Bland-Altman analysis to estimate the agreement between the methods. Moreover, we calculated mean baseline and mean fit for 50 one-month old rats to identify potential systematic errors in fits. Finally, results from an exemplary experiment obtained with LCModelTM were reproduced with TARQUIN. Bland Altman plots indicate that there is an acceptable agreement between LCModelTM and Tarquin with adjacent basis set for total N-Acetylo-aspartate (tNAA), total-choline (tCho), total-creatine (tCr) and glutamine/glutamate (Glx) (95% confidence interval of agreement below 20%). However, for both basis sets, Tarquin gave significantly more variable results in myo-Inositol comparing to LCModel. In conclusion, despite some potential biases to the results, spectra were successfully processed with Tarquin and they yielded similar results to those obtained with LCModel.
1H-MRS, quantification, basis set, comparison with LCModel, absolute concentrations, relative concentrations 7 < 8
We have recently observed that one-year use of high fat diet (HFD) that induced mild ketonemia lead to better learning and memory, larger hippocampi volumes without any changes to cortical volumes, as well as higher concentrations of total NAA (tNAA: N-acetylaspartate and N-acetylaspartateglutame; marker of neuronal viability), total Cho (tCho: Glycerophosphocholine +Phosphocholine, which are believed to be primarily involved in cell membrane breakdown and synthesis) and total Cr (tCr: creatine + phospo-creatine – involved in cell bioenergetics){Setkowicz, 2015 #1108} We performed ROI analyses and used LC Model for spectral processing.
However, the spectroscopic results may have been driven by specific processing procedures used by LC Model, thus we needed to use a different software to assure the obtained results are independent of processing procedure. TARQUIN (Wilson et al, 2011) [1] is an open source alternative that was demonstrated to work comparably well to LCModelTM with wide range of 1.5T and 3.0T proton spectra.
However, it has not been used to fit proton, animal spectra acquired at 7.0T.
There are several metabolite quantification programs available for research (and commercial) use. They differ in procedures, assumptions about the line shapes, baseline, macromolecule and lipid contributions, as well as in soft-constrains. Thus, a direct comparison between these programs is not straightforward or even not possible. The code of some programs is not publicly available.
Currently, the most widely used procedure is LC Model™ [2]. It is one of the first algorithms to incorporate a metabolite basis set into the fitting model and is widely used for the analysis of short-echo time 1H MRS data. However, the costs of acquiring this software is substantial. There is a lot of interest in use of alternative packages to process spectroscopic data, such as Tarquin. Its comparable quality of spectral processing with LC Model was demonstrated using a wide range of spectra acquired on 1.5T and 3T scanners, as well as using Monte-Carlo simulations [1]. However, the procedures of processing data obtained at 7T systems have not been established.
Direct comparison between TARQUIN and LC Model is not straightforward, given the known and unknown differences in processing (LC Model is a commercial program, thus its code is not open to public). The major differences are as follows: TARQUIN uses a time-domain fitting combined with algorithmic approach taken from AQSES [3]. It deletes starting points from the free induction decay (FID) to eliminate signal-baseline interference. Basis set is synthesized from simulated metabolites signal, lipids and macromolecules. To estimate the amplitudes TARQUIN utilizes a non-negative least squares projection. LC Model models data in the frequency domain using a linear combination of metabolite, lipid and macro-molecule signals combined with a smoothing splines to account for baseline signals. Both programs use different fitting algorithms: TARQUIN uses Levenberg-Marquardt algorithm with the variable projection method due to reduction of model parameters, whereas LC Model utilizes Marquard modification of a constrained Gauss-Newton least squares method. To fit baseline, TARQUIN smooths residual signal with a convolution filter, whereas LCModel uses cubic b-splines for fitting. Programs use different sets of soft constrains. LC Model additionally uses aspartate, GABA, glucose, scyllo-Inositol, in addition to NAA/NAAG ratio used by both programs.
We have proposed two basis-sets for TARQUIN to work with 7T spectra (described in the Methods section), processed 7T hippocampal and anterior cingulate cortex (ACC) spectra obtained in a group of 50 one-month old Wistar rats, as well as again in 47 of them when they reached one year of age. Then we compared these fittings with LCModel fittings of the same spectra. Given the intrinsic differences between both fitting procedures, we evaluated quality of fit (Q) and performed Bland-Altman analysis to estimate the agreement between these two methods. Moreover, we calculated mean baseline and mean fit for 50 rats at one month of age. In order to visually estimate biased in spectral fitting. Finally, results from an exemplary experiment obtained with LCModelTM were reproduced and extended by use of TARQUIN.
Animals
The spectra evaluated in this study were acquired as part of another project evaluating the long-term effects of high-fat diet (HFD) on memory and memory-related regions in the brains of Wistar rats. The animals were scanned at one month, before half of them were switched to HFD, and again at one year of age. Blood level of sugar in the group fed with HFD was 6.5% higher than in the control group (p=0.01), but still within normal values. The details are described elsewhere [4]. All procedures involving the use of animals were approved by the Bioethical Commission of the Jagiellonian University in Krakow, Poland in accordance with international standards.
Spectroscopic data acquisition
At first month and at the 12th month of age, brains of the examined animals were scanned with 7T Bruker BioSpec 70/30 Avance III, with a quadrature volume coil (15 cm inner diameter) and a phased array receiver (2x2 elements) positioned over the animal’s head. The receiver coil position was adjusted to obtain high signal intensity over regions of interest. The animals were positioned prone with the head placed in the stereotactic apparatus and anesthesia mask, and were anesthetized with 1.5% isoflurane in a mixture of oxygen and air. Respiration, heart rate, and oxygen saturation were monitored throughout the experiment. Rectal temperature was kept at 37OC by placing the animal on top of temperature controlled warm water blanket. Tripilot scans were used for accurate positioning of the animals inside the magnet. Linear and second order global shims were adjusted with ADJ_1st_2nd_order protocol. Afterwards, linear and second order local shims were automatically adjusted with FASTMAP in a cubic volume which contained the volume of interest region. The unsuppressed water line width was typically maintained at around 10-15 Hz. Spectra were obtained by localized proton spectroscopy at short echo using PRESS sequence (TR/TE = 3500/20 ms, 256 averages, 8,192 points, TA=15min) with VAPOR water suppression, the outer volume suppression, and frequency drift correction (flip angle 7 deg.). Each measurement was carried out in a two volumes of interest: 8 x 2 x 2 mm encompassing hippocampus and 4 x 4 x 2 mm volume in ACC (Figure 1).
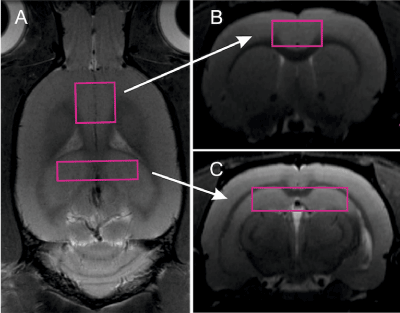
Figure 1. Locations of volumes of interest: voxel including hippocampus (a), voxel including anterior cingulate cortex, used as control in the study.
All spectra were quantified using: 1) with TARQUIN a constrained least-squares approach to the automated quantization of in-vivo 1H magnetic resonance spectroscopy data [1] and 2) a linear combination analysis method LCModel (Stephen Provencher Inc, Oakville, Ontario, Canada; [2] including macromolecule spectrum in the database and priors provided by the manufacturer. In the former case we used two sets of macromolecular spectra described in the following paragraph. The unsuppressed water signal measured from the same volume of interest was used for eddy-currents corrections and as internal reference for absolute metabolite quantification. Metabolite concentrations are reported in institutional units (i.u.).
Spectral fitting with Tarquin
To allow for meaningful comparisons, we selected the same set of metabolites that is used in LCModel fitting. For TARQUIN, we evaluated two macromolecular basis sets for: 1) based on LCModel (to allow more direct comparison with LC Model), further reffered to as LCModel-like basis set and 2) based on work by Hong and colleagues [5], further reffered to as Hong-like basis set; it consists of 16 macromolecules. Both basis sets are included in supplementary material. We experimentally adjusted the starting point parameter to be 20 and the end point to be 2000 (at least twice long as visible metabolite signal); we used default value of λ= 0.2 (default soft-constraints influence). All processed spectra were visually reviewed for quality. Only metabolites that had demonstrated good reliability on test-retest [6] were used in analyses: tNAA (N-Acetylaspartate + N-Acetylaspartylglutamate), tCr (Creatine + Phosphocreatine), tCho (Glycerophosphocholine +Phosphocholine), Ins (myo-Inositol), GABA (γ-Aminobutyric Acid), and Tau (Taurine).
Statistical analyses
Quality of the fit and Bland Altman analysis were done on the complete dataset of all 194 spectra acquired in the study. Subsequent effect size compartment was done on selected groups detailed in each section description.
Quality of the fit
Quality of the fit (Q) was defined as the standard deviation of the residual signal between 0.2 and 4.0 ppm divided by the standard deviation of the spectral noise [1]. Spectral noise parameter was estimated according to [7], i.e., residual signal was divided into 31 equally sized intervals and for each interval the standard deviation (SD) of the signal was calculated; the interval with the smallest SD was the used as the noise estimator. According to the definition [7], Q will be: 1) less than unity where overfitting has occurred and 2) greater than unity when the signal has not been completely modeled. Q cannot be used to identify baseline problems or artifacts. To ensure well spectra quality all datasets were visually inspected. Two heavily corrupted spectra were removed from analyses.
Bland Altman analysis
Bland Altman analyses were performed separately for concentration of tNAA, Glx, Ins, tCho, tCr, to evaluate agreement between the TARQUIN and LCModel [8], independently for each basis set. The average of the upper and lower limits of agreement, which represent the 95% confidence intervals of agreement, were calculated for each metabolite to estimate the agreement between LC Model and TARQUIN. The first step is to examine the data. A simple plot of the results of one method against those of the other. Usually all the data points will be clustered near the line. A plot of the difference between the methods against their mean provides information on the agreement. It is assumed that this differences should be less than 20%. Reproducibility coefficients, which are equivalent to 95% confidence intervals of agreement, represent numerically the agreement between the fitting methods.
Evaluation of mean baselines
For each of the processing method, all spectra were aligned and added up to evaluate for potential biases in fitting.
An exemplary study
In this paragraph, differences between treatment and control groups obtained with different postprocessing methodology were exemplified. Hippocampal spectra were selected. Groups were compared in seven metabolites including tNAA, tCr, tCho, Glx, Ins and Glc (glucose). 25 spectra were obtained in the group on HFD, whereas the remaining 22 spectra in the control group.
Quality of the fit
Quality of the fit (Q) in 194 spectra dataset (both VOIs, both time-points) was: 1.17 (SD=0.08) for LCModel, 1.27 (SD=0.13) for TARQUIN using the LCModel-like basis set, and 1.20 (SD=0.10) for TARQUIN with Hong-like basis set. Overfitting (Q<1) did not occur for any of the analyzed spectra (Figure 2).
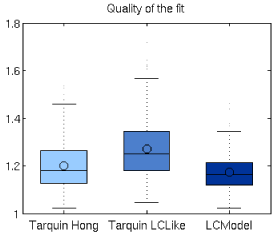
Figure 2. Quality of fit (Q) for LCModel and TARQUIN-fittings with Hong-like and LCModel-like basis sets. Q>1 is consistent with the fact that spectral signal was not completely modeled in any case.
Bland-Altmann analyses
Bland Altman plots indicate that there is an acceptable agreement between LCModel and Tarquin with LCModel-like basis sets for tNAA, tCho, tCr and Glx (95% confidence interval of agreement below 20%), but not for myo-Inositol (>28%, i.e., 95% confidence interval of agreement above 28%). However, when comparing LCModel with TARQUIN utilizing Hong-like basis set, significantly higher disagreement in the results can be observed, i.e. tCh (35 %), tCr (24%). The good agreement between both TARQUIN methods, together with lack of agreement between LCModelTM and TARQUIN, seems to point to differences in residual water peak fitting between TARQUIN and LCModelTM. All results are presented in Table 1. Exemplary plots for Blant-Altman analyses for NAA + NAAG are presented in Figure 3.
Table 1. Results
|
LCModel vs TARQUIN HONG |
LCModel vs TARQUIN LCLike |
HONG vs LCLike |
tNAA |
18% |
17% |
12% |
tCho |
35% |
18% |
36% |
tCr |
24% |
17% |
17% |
Glx |
19% |
17% |
18% |
Ins |
31% |
28% |
18% |
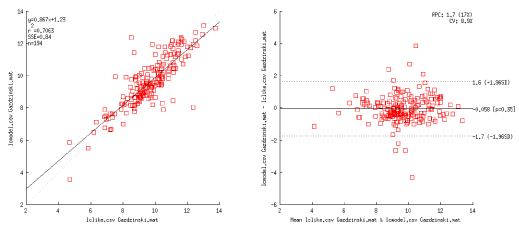
Figure 3. Representative results for Blant-Altman analysis, here for NAA+NAAG for TARQUIN (LCModel-like basis-set) vs LCModel.
As expected, TARQUIN utilizing LCModel-like basis set shows better agreement with LCModelTM, than TARQUIN using Hong-like basis set.
Mean baselines
This paragraph compares fittings and potential systematic errors for both fittings with TARQUIN and with LCModelTM. Figure 4, 5 and 6 presents average fit, average baseline, and averaged residuals for a) TARQUIN with LCModel-like basis set, b) TARQUIN with Hong-like basis set, and c) LCModel for 194 analyzed spectra.
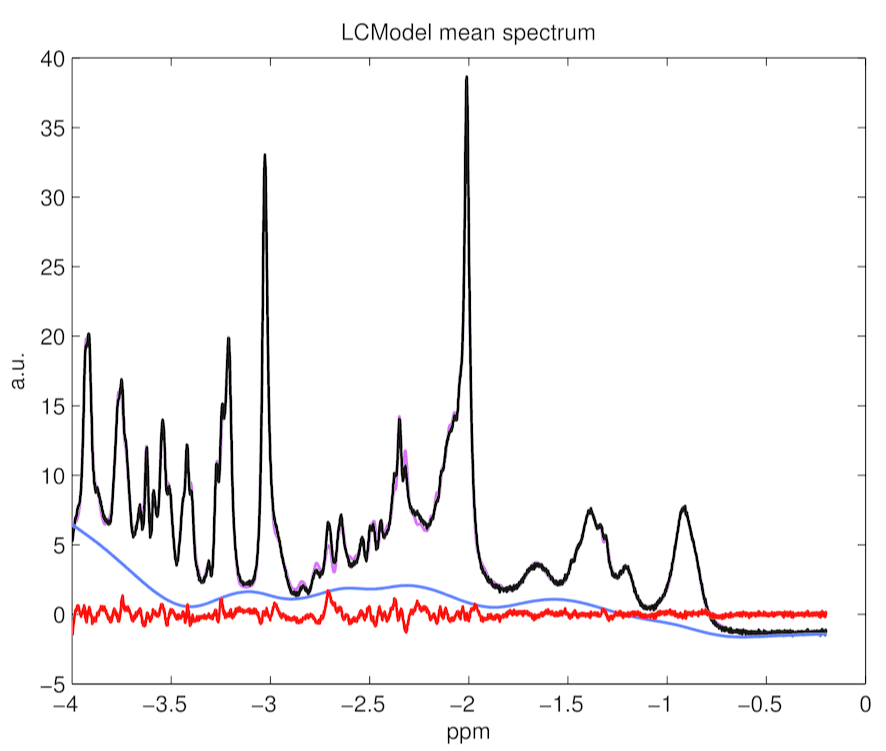
Figure 4. LCModel mean spectrum, baseline, residuals and fitting estimates for 194 rat spectra.
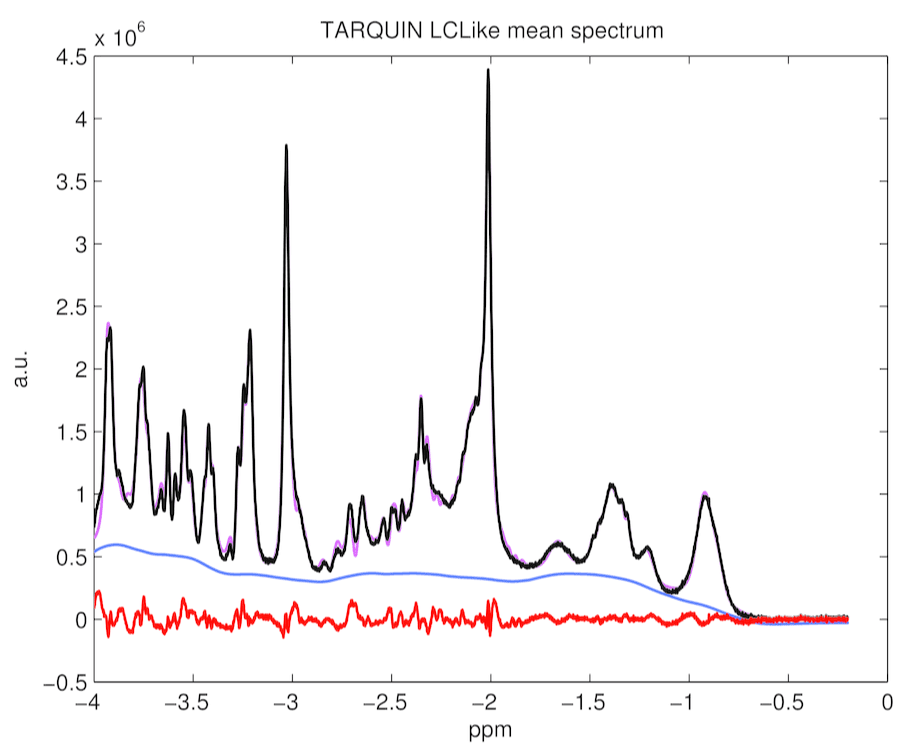
Figure 5. TARQUIN with LCModel-like basis set mean spectrum, baseline, residuals and fitting estimates for 194 rat spectra. It appears lots of systematic errors in fitting are present.
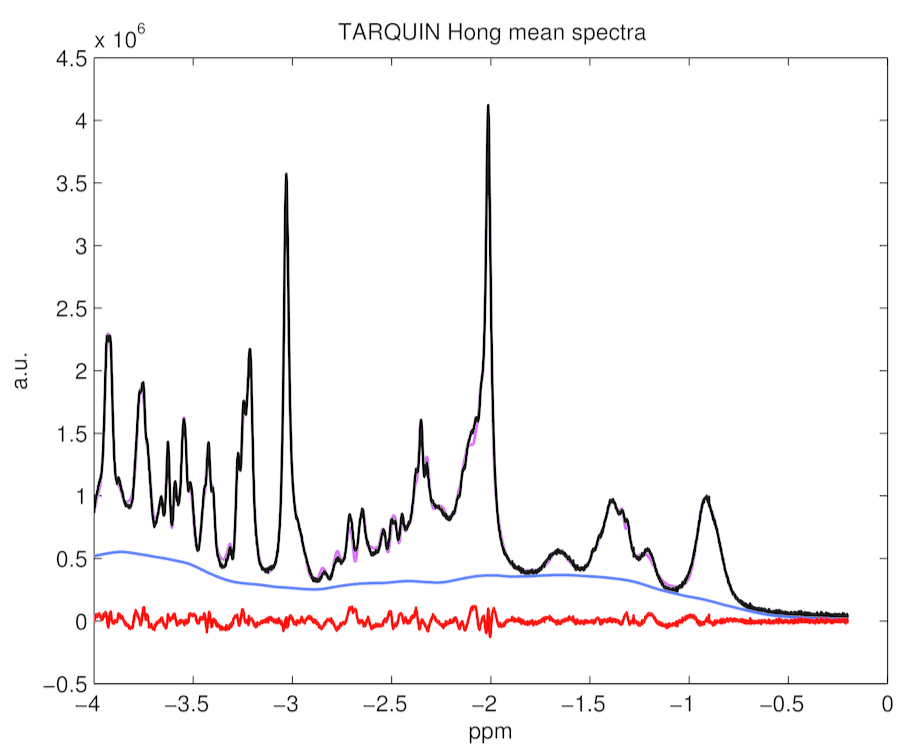
Figure 6. TARQUIN with Hong basis set mean spectrum, baseline, residuals and fitting estimates for 194 rat spectra
An exemplary study
The concentration of tNAA was higher by 10.4% (p=0.01), Glx by 6% (p=0.07) and tCr by 8.5% (p=0.003). There was also a significant change in Glc level of 19% (p=0.015), larger than the 6.5% difference observed via blood panels. On the other hand, TARQUIN, indicate differences in tNAA, Glx and tCr respectively (6.8%, p=0.01, 6.5%, p=0.03, and 4.5%, p=0.03, respectively), consistent with the results obtained with LCModelTM (Figure 7 and 8).
2021 Copyright OAT. All rights reserv
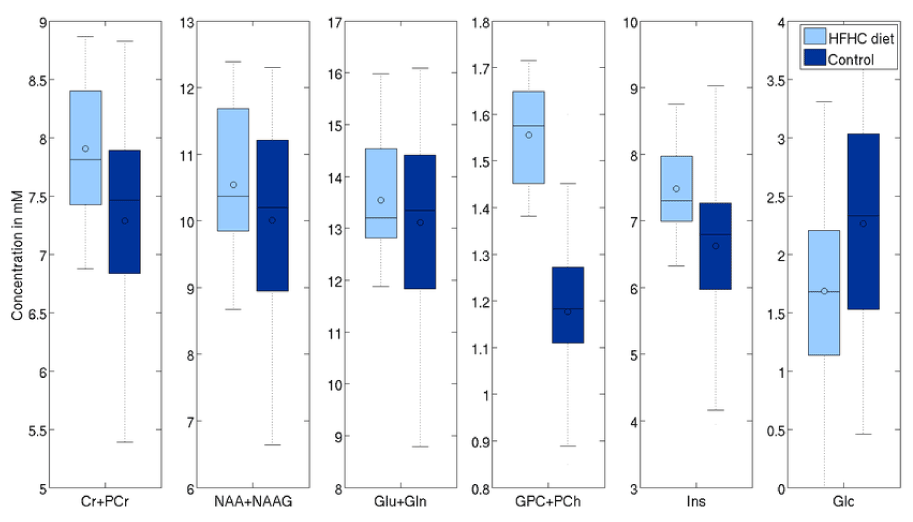
Figure 7. Effects of aging on selected metabolites (processed with LCModel).
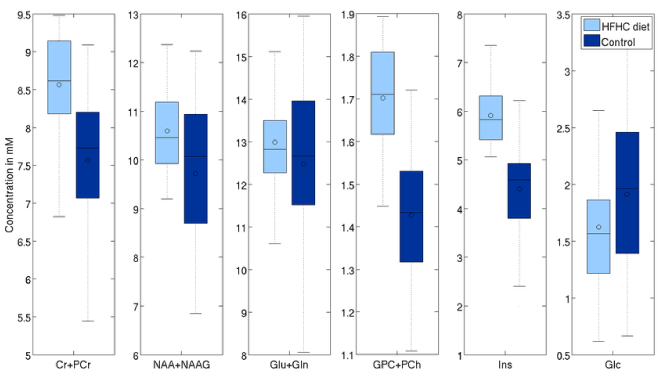
Figure 8. Effects of aging on selected metabolites (processed with TARQUIN with LCModel-like basis set).
Second analysis was done on the 22 one-year old vs 50 nursling rats. Differences with significance level (uncorrected) p<0.05 are visible in tCr (4.7% p<0.01), tCho (33.2% p<0.01), Ins (7.5% p<0.01) and Glc (-28% p=0.03) in LCModel. On the other hand, TARQUIN reports tCr (11.5% p<0.01), tCho (19.3% p<0.01), Ins (26.8% p<0.01) and Glc by -20% with p=0.05 and, what is more comparing to LCModel, tNAA (3.84% p=0.02) (Figure 9 and 10).
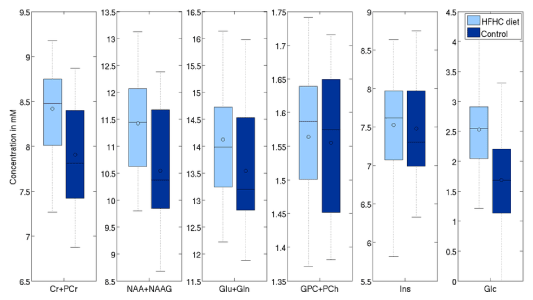
Figure 9. Effects of HFD on selected metabolites (processed with LCModelTM).
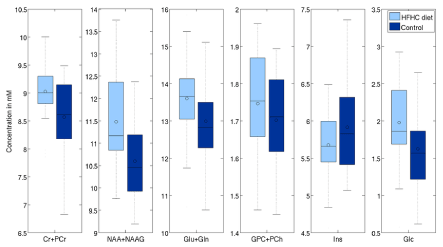
Figure 10. Effects of HFD on selected metabolites (processed with TARQUIN using the LCModel-like basis set).
Our results confirm that application of TARQUIN may be extend to 7T animal spectra and yield results comparable with LCModelTM for basis sets adapted from other work.
TARQUIN was previously demonstrated to robustly fit clinical spectra obtained at 1.5T and 3T in a broad spectrum of participants ranging from healthy volunteers to patients suffering for cancers [1].
Results obtained with Hong-like basis set performs better than LCLike due to the wider set of macromolecules. It is the matter of further comparisons to decide, whether the reacher basis set, helps or makes the fitting too complicated to give stable metabolites results.
The limitation of our analyses is due to adaptation of existing basis sets. We strongly believe that work is needed to establish basis sets for 7T data.
In conclusion, despite some potential biases to the results, spectra were successfully processed with Tarquin and they yielded similar results to those obtained with LCModel. More work is needed to adapt Tarquin to fit 7T spectroscopic data. Creating a macromolecular basis set at 7T appears to be a must.
This project was supported by Polish National Science Centre (grant 2011/03/B/NZ4/03771 to SPG).
- Wilson M, Reynolds G, Kauppinen RA, Arvanitis TN, Peet AC (2011) A Constrained Least-Squares Approach to the Automated Quantitation of In Vivo H-1 Magnetic Resonance Spectroscopy Data. Magnetic Resonance in Medicine 65: 1-12.
- Provencher SW (2001) Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed 14: 260-264. [Crossref]
- Poullet J-B, Sima DM, Simonetti AW, De Neuter B, Vanhamme L, et al. (2007) An automated quantitation of short echo time MRS spectra in an open source software environment: AQSES. NMR Biomed 20: 493-504. [Crossref]
- Gazdzinski S, Setkowicz Z, Osoba J, Karwowska K, Majka P, et al. (2014) Is high-fat high-carbohydrate diet (HFCD) neuroprotective? A magnetic resonance imaging study in Wistar rats. European Journal of Neurology 21: 391-391.
- Hong ST, Pohmann R (2013) Quantification issues of in vivo (1) H NMR spectroscopy of the rat brain investigated at 16.4 T. NMR Biomed 26: 74-82. [Crossref]
- Wijtenburg SA, Rowland LM, Edden RAE, Barker PB (2013) Reproducibility of brain spectroscopy at 7T using conventional localization and spectral editing techniques. J Magn Reson Imaging 38: 460-467. [Crossref]
- Slotboom J, Nirkko A, Brekenfeld C, van Ormondt D (2009) Reliability testing of in vivo magnetic resonance spectroscopy (MRS) signals and signal artifact reduction by order statistic filtering. Measurement Science & Technology 20(10).
- Bland JM, Altman DG (2010) Statistical methods for assessing agreement between two methods of clinical measurement. Int J Nurs Stud. 47: 931-936.










