Type 2 diabetes mellitus (T2DM) is a chronic metabolic disease characterized by hyperglycemia commonly associated with insulin resistance at high risk of renal, neurological and cardiovascular complications. It is defined as a multifactorial etiology disease where genetic predisposition and lifestyle play an important role in pathophysiology and onset. Recently, genome wide association studies (GWAS) have been widely used to identify deregulated expression of T2DM related genes and genetic risk factors that can contribute, together with environmental and behavior factors, to T2DM onset. Since its dual feature, anti-diabetic effective therapy need to acknowledge the genetic contribution to T2DM pathophysiology. The pharmacological treatment of T2DM depends on blood glucose levels and/or glycated hemoglobin (HbAc1): well-compensated patients with normal HbA1c levels are, generally, treated with oral hypoglycemic drugs, such as metformin, associated with a diet that limits carbohydrate intake. Conversely, the uncompensated patient, with high levels of HbAc1 is generally treated with insulin or other new generation drugs or a combination of them. Given the multifactorial nature of T2DM, recent studies have identified personalized therapy as a powerful means to refine the effectiveness of the therapy itself, paving the way for precision medicine.
In the last decade, Type 2 diabetes mellitus (T2DM) incidence have dramatically grown up worldwide [1], with serious consequences to global health. T2DM is a multifactorial disease where metabolic impairment is the common feature: it is clinically characterized by high blood glucose levels that can lead to an inadequate insulin secretion, insulin resistance and/or glucagon secretion impairment, often associated with comorbidity [2]. According to the ADA (American Diabetes Association) guidelines, T2DM diagnosis is based on plasma glucose criteria, either the fasting plasma glucose (FPG) value, or the 2-hour plasma glucose (2-h PG) value during a 75-g oral glucose tolerance test (OGTT), or glycated hemoglobin (HbA1c) percentage evaluation [3]. In particular, T2DM diagnosis requests FPG ≥ 126 mg/dl, OGTT≥ 200mg/dl and HbA1c ≥ 6.5% [3]. Pharmacological therapy is combined taking in account both FPG and HbA1c; moreover, ADA guidelines recommend life style modification coupled with drugs therapy depending on HbA1c percentage and/or body mass index levels: HbA1c levels less of 6.5%, in fact, requested changing in lifestyle coupled with only moderate metformin administration [4]. On the other hand, for HbA1c levels less than 7% therapy based on sodium-glucose transport protein 2 (SGLT-2) inhibitors, Dipeptidyl peptidase-4 (DPP-IV), or glucagon-like peptide 1-receptor agonist (GLP1-RA) may be suggested [4] while highly uncompensated T2DM patients showing HbA1c percentage from 8 up to 9%, need insulin treatment alone or in combination with other hypoglycemic drugs [4]. Despite the introduction of many novel anti diabetic drugs, such as gliflozin, whit promising results especially on cardiovascular complication preventions [5], or renal failure outcomes amelioration [6], non-responder patients to therapies are a serious challenge to face with [7]. A recent study highlighted how a significant percentage of newly diagnosed patients resulted as metformin treatment non-responder [8]. Moreover, Heerspink and colleagues demonstrated how Dapagliflozin treatment failed to decrease urinary albumin-to-creatinine ratio in 46% of clinical cases [9]. Based on all these findings, a genomic approach could be useful to develop novel and more personalized therapies to treat the non-responders T2DM patients [10].
The genetic component that characterizes the etiology of T2DM must be taken into consideration in developing the therapeutic plan for patients. Studies conducted on the genetics of T2DM using the genome-wide association studies (GWAS) technique, identified about 400 genetic loci strongly associated with T2DM [11]. GWAS is an observational study aimed to identify genetic variants across whole genome that can be related to a certain disease; it was developed in 2002 and recently became an important tool of genetics research [12,13]. GWAS analysis can be also used to determine the risk of developing disease [14,15]. Moreover, this technique identifies single nucleotide polymorphism (SNPs) that are associated with a large number of disorders [16]. Thirty-eight SNPs tightly associated with T2DM have been initially found [17], together with several SNPs linked with hyperglycemic state and high triglyceride conditions [18]. Nowadays, GWAS identified at least 143 genetic variants associated with T2DM [19]; moreover, in human islets 2,949 SNPs associated with in vivo cis-regulatory elements have been recently identified [20]. GWAS technique could potentially leading to discover many new pharmacological targets for T2DM therapy. Recently polymorphisms in SLC30A8 as affecting disease risk has been discovered in T2DM [21]. Thus, the common polymorphism rs13266634, associated with increased risk for developing T2DM, was associated with lowered beta cell function and a 14% increase in diabetes abundance [21]; it seems clear that a patient with this particular genetic setup will need a more precise therapeutic approach compared to a wild type genetic setup patient. Pharmacogenetic study on T2DM highlighted as mutations in KCNJ11 gene encoding for ATP (Adenosine Triphosphate) sensitive potassium channel subunit Kir 6.2, reduces insulin secretion causing permanent neonatal diabetes onset; thus, patients with this mutation have more benefits in sulfonylureas therapy instead insulin [22].
Precision medicine is a customized approach that allows coupling patient whit the best therapy taking in account his/her genetic profile environments and lifestyles leading to a personalized pharmacological intervention [23]. To achieve this goal, as previously reported, the genetic profile of diabetic subjects plays a pivotal role in avoiding ineffective therapies. In agreement, the GoDARTS (Genetics of Diabetes Audit and Research in Tayside Scotland) study demonstrated how the reduced allelic functions of the OCT1 gene, encoding the organic cationic co-trasporter –OCT, is directly correlated with the increased intolerance to metformin [24]. The rationale behind precision medicine is avoiding administration of an ineffective therapy that could not bring any benefit to the patient (Figure 1).
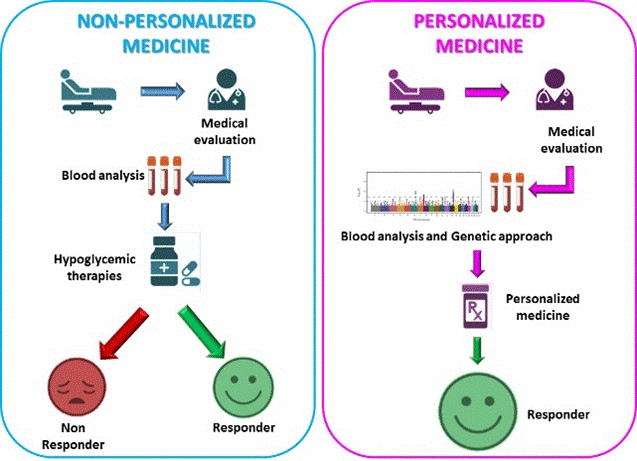
Figure 1. Schematic representation of personalized medicine rationale
Moreover, it has also been suggested that the low allelic expression of the channels responsible for serotonin retention, is specifically correlated with the increased development of intolerance to metformin [25]. Based on these studies, metformin therapy would be contraindicated in patients who presented a genetic profile that included the reduced expression of the above mentioned alleles. Another study established as the presence of genetic polymorphisms, particularly TCF7L2 polymorphism, could influence the response to DPP-IV inhibitors in diabetic patient [26]. Thus, antidiabetic therapy involving the use of DPP-IV inhibitors for T2DM patients with TCF7L2 polymorphism, would invalidate or make less effective the prescribed therapy. Moreover, published data demonstrated the presence of genetic polymorphisms able to reduce the response to GLP-1-RA inducing T2DM [27]. This study confirmed uselessness to administrate GLP-1-RA therapy to a non-responder patient.
Precision medicine associated with T2DM therapy is becoming the future prospect for the development of a strategy that takes into account the patient's genetic cluster. Further studies aimed to identify new genetic targets capable of influencing the response or tolerance to an antidiabetic drug are mandatory to develop a personalized approach, in order to avoid prescribing drugs whose efficacy may not be effective. Physicians and diabetologists need to work in synergy with geneticists in order to elaborate a more effective therapeutic approach that could be as personalized as possible.
- Geiss LS, Wang J, Cheng YJ, Thompson TJ, Barker L, et al., (2014) Prevalence and incidence trends for diagnosed diabetes among adults aged 20 to 79 years, United States, 1980-2012. JAMA 312: 1218-1226. [Crossref]
- Smyth S, Heron A (2006) Diabetes and obesity: the twin epidemics. Nat Med 12: 75-80. [Crossref]
- American Diabetes Association (2020) 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2020. Diabetes Care 2020. 43: S14-S31. [Crossref]
- American Diabetes Association (2021) 3. Prevention or Delay of Type 2 Diabetes: Standards of Medical Care in Diabetes-2021. Diabetes Care 44: S34-S39. [Crossref]
- Akinci B (2019) Dapagliflozin and Cardiovascular Outcomes in Type 2 Diabetes. N Engl J Med 380: 1881. [Crossref]
- Perkovic V, Jardine MJ, Neal B, Bompoint S, Heerspink HJL, et al., (2019) Canagliflozin and Renal Outcomes in Type 2 Diabetes and Nephropathy. N Engl J Med 380: 2295-2306. [Crossref]
- Kim SA, Shim WH, Lee EH, Lee YM, Hee Beom S, et al., (2011) Predictive clinical parameters for the therapeutic efficacy of sitagliptin in korean type 2 diabetes mellitus. Diabetes Metab J 35: 159-165. [Crossref]
- Rashid M, Shahzad M, Mahmood S, Khan K (2019) Variability in the therapeutic response of Metformin treatment in patients with type 2 diabetes mellitus. Pak J Med Sci 35: 71-76. [Crossref]
- Heerspink HJL, David Sjöström C, Inzucchi SE, Hallow MK, Cain VA, et al., (2019) Reduction in albuminuria with dapagliflozin cannot be predicted by baseline clinical characteristics or changes in most other risk markers. Diabetes Obes Metab 21: 720-725. [Crossref]
- Jameson JL, Longo DL (2015) Precision medicine--personalized, problematic, and promising. N Engl J Med 372: 2229-2234. [Crossref]
- Mahajan A, Wessel J, Willems SM, Zhao W, Robertson RN, et al., (2018) Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet 50: 559-571. [Crossref]
- Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, et al., (2002) Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet 32: 650-654. [Crossref]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, et al., (2005) Complement factor H polymorphism in age-related macular degeneration. Science 308: 385-389. [Crossref]
- Ingram VM (1956) A specific chemical difference between the globins of normal human and sickle-cell anaemia haemoglobin. Nature 178: 792-794. [Crossref]
- Chang JC, Kan YW (1979) Beta 0 thalassemia, a nonsense mutation in man. Proc Natl Acad Sci U S A 76: 2886-2889. [Crossref]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928-933.
- Billings LK, Florez JC (2010) The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci 1212: 59-77. [Crossref]
- Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331-1336. [Crossref]
- Xue A, Wu Y, Zhu Z, Zhang F, Kemper KE, et al., (2018) Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat Commun 9: 2941. [Crossref]
- Khetan S, Kursawe R, Youn A, Lawlor N, Jillette A, et al., (2018) Type 2 Diabetes-Associated Genetic Variants Regulate Chromatin Accessibility in Human Islets. Diabetes 67: 2466-2477. [Crossref]
- Rutter GA, Chimienti F (2015) SLC30A8 mutations in type 2 diabetes. Diabetologia 58: 31-36. [Crossref]
- Pearson ER, Flechtner I, Njølstad PR, Malecki MT, Flanagan SE, et al., (2006) Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N Engl J Med 355: 467-477. [Crossref]
- National Research Council (2011) Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington (DC). [Crossref]
- Zhou K, Donnelly LA, Kimber CH, Donnan PT, Doney ASF, et al., (2009) Reduced-function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes 58: 1434-1439. [Crossref]
- Mahrooz A, Parsanasab H, Hashemi-Soteh MB, Kashi Z, Bahar A, et al., (2015) The role of clinical response to metformin in patients newly diagnosed with type 2 diabetes: a monotherapy study. Clin Exp Med 15: 159-165. [Crossref]
- Zimdahl H, Ittrich C, Graefe-Mody U, Boehm BO, Mark M, et al., (2014) Influence of TCF7L2 gene variants on the therapeutic response to the dipeptidylpeptidase-4 inhibitor linagliptin. Diabetologia 57: 1869-1875. [Crossref]
- Hart LM’t, Fritsche A, Nijpels G, Leeuwen NV, Donnelly LA, et al., (2013) The CTRB1/2 locus affects diabetes susceptibility and treatment via the incretin pathway. Diabetes 62: 3275-3281. [Crossref]

