Fetal to neonatal transition of heart is an elaborate process, during which neonatal cardiomyocytes undergo functional maturation and terminal exit from the cell cycle. This tightly regulated process may become rapidly disrupted in the context of congenital heart defects (CHDs). CHDs affect 1% of live births and are a major source of childhood morbidity and mortality. Residing in the non-coding regions of the genome, the long noncoding RNAs (lncRNAs) are increasingly recognized as important regulators of cardiac development and putative key contributors to CHDs. However, lacking sequence conservation and appropriate annotation, elucidating lncRNAs functions has been notoriously challenging. While the roles of most predicted lncRNAs in heart development and CHDs await functional resolution, the biological impacts of regulatory lncRNAs and their mechanisms of action are likely to be diverse, thus requiring focused efforts in these contexts. These limitations can now be circumvented by the combination of advanced, genome-wide, sequencing platforms and powerful functional and molecular tools. In this review article, we discuss principle considerations for regulatory lncRNA discovery. Although we primarily focus on neonatal heart maturation and CHDs, the proposed algorithm should facilitate mechanistic exploration of functional lncRNAs in various disease models. These insights could ultimately lead to novel diagnostics and therapeutic approaches targeting lncRNAs in neonatal heart pathologies and CHDs.
Long noncoding RNA; Congenital Heart Defects; Genome; Epigenome; Transcriptome; Neonatal Heart Maturation.
Revealed as a mysterious layer between the genome and the proteome, the long noncoding RNAs have revolutionized the traditional view of central dogma of biology by shedding lights to the dark matter of the genome and leading to exciting opportunities for further understanding of developmental biology and human diseases as putative disease modifiers, diagnostic biomarkers, and therapeutic targets.
The finding that most of the genome in complex organisms is transcribed has significantly shifted our understanding of genomic and cellular biology. Parallel to current advancement of genome-wide transcriptome characterization, by implementing deep RNA-sequencing and advanced bioinformatics analysis tools [1-4], the recent discoveries of the regulatory lncRNAs, which exert diverse functions and can potentially be implicated in development and disease have led to growing interest in deciphering their functional roles in mammalian biology and human disease [1-6]. In particular, there is an increasing interest in investigating their roles in heart development and disease [7-11]. Several reports have identified thousands of lncRNAs enriched in heart, dynamically transcribed during cardiac development and differentially regulated in cardiovascular diseases [9-12]. For example, Bvht (Braveheart) was found to regulate the core cardiac transcription network during cardiogenesis, acting in trans by interacting with SUZ12, a core component of the polycomb repressive complex 2(PRC2) [8], and MIAT (myocardial infarction-associated transcript) was identified by genome-wide association studies GWAS as a risk factor for myocardial infarction [12]. Nevertheless, the precise mechanistic details on how lncRNAs expression may affect heart development or induce heart pathology remain limited to few reports and we are only at the beginning to understand the putative implications of these emerging genomic regulators in CHDs. Developmental and functional maturation of neonatal heart is determined by its gene expression network, which is regulated by genetic and epigenetic mechanisms, including lncRNAs [1-4]. The proved links between lncRNAs and heart development [8-10] indicate that lncRNAs comprise core transcriptional regulatory circuits with key transcription factors. Furthermore, lncRNAs can potentially mediate pathological response to hemodynamic and environmental stress factors in the context of CHDs. Therefore, establishing lncRNAs function can yield novel biomarkers and targets for therapy. Elucidating the functional roles of these emerging regulators is a crucial step toward important translational implications in neonatal heart maturation in the context of CHDs.
First discovered in the early 1990s, lncRNAs are arbitrarily defined as the RNA species of >200 nucleotides in length, lacking a significant open reading frame (ORF), and much less conserved than the protein-coding genes [9-11]. As a transcriptional class, the lncRNAs are grouped according to their genomic location and orientation as compared to the closest coding gene [13] (Figure 1). Intergenic lncRNAs are located in between two coding genes, not overlapping therefore with any coding sequences. A subclass of intergenic lncRNA is enhancer-associated lncRNAs, many of which are bidirectional, lacking a poly A tail, and expressed at low copy number. In contrast, intragenic lncRNAs overlap with coding sequences. In this case, lncRNAs can be transcribed from the sense or the antisense strand. A subclass of intragenic overlapping antisense lncRNAs is the natural antisense transcripts (NATs), which complement and regulate their opposite coding genes expression. Intronic lncRNAs represent a class of lncRNAs that are encoded within introns of protein-coding genes. Finally, bidirectional lncRNAs are transcribed at the close vicinity of a protein-coding gene on the opposite strand [5,6,17-20].
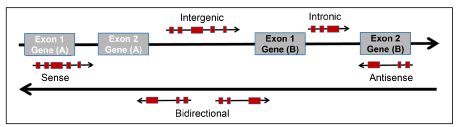
Figure 1. Genomic Position of LncRNAs. Protein-coding genes and their exons are represented in grey, while lncRNAs and their exons are represented in red. Intergenic lncRNA, transcribed intergenically from one or both strands (Bidirectional). Intronic lncRNA, transcribed entirely from introns of protein-coding genes. Sense lncRNA, transcribed from the sense strand of protein-coding genes, overlapping with protein-coding sequence. Antisense lncRNA, transcribed from the antisense strand of protein-coding genes, overlapping with protein-coding sequence.
LncRNAs are pervasively transcribed throughout the genome displaying remarkable similarities to classical messenger RNAs (mRNAs) in that they are transcribed by RNA Polymerase II (RNAPII), 5’ capped, and polyA tailed, and are generally, alternatively spliced [18-22]. Compared to mRNAs, most lncRNAs are expressed at relatively lower levels, associating with chromatin modifying complexes or splicing machinery. Although some lncRNAs are expressed at levels comparable to mRNAs, those appear to function as structural scaffolds for nuclear domains. Finally, lncRNAs may exhibit specific subcellular localization (cytoplasmic vs nucleus), suggesting roles in regulating specialized cellular functions in these compartments [23].
Featuring temporal regulation, and tissue and cell specificity, as well as functional diversity [24-26], the lncRNAs have increasingly expanded the functional complexity of transcriptome. It is increasingly evident that lncRNAs play important roles in regulating gene expression at different levels, including epigenetic modification, transcription regulation, and post‐ transcriptional processing [24]. While the natural antisense transcripts (NATs) are most likely to repress expression of their complementary counterpart [17, 20], which in most cases lies directly in the opposite strand at the same genomic locus, the other classes, however, have diverse functions derived from their ability to fold into complex secondary and tertiary structures. Furthermore, lncRNAs can partially hybridize with DNA leading to the formation of scaffold structures for RNA processing, histone modification, protein binding, and to a minimal extent, protein coding. Moreover, lncRNAs can interact with proteins, modifying their activities, dictating their localizations, and executing their functions via acting as decoys, scaffolds or guides. Taken together, unlike microRNAs, which uniformly carry out repressive functions, lncRNAs are thought to exert diverse regulatory potential acting in cis or in trans to induce or suppress the expression of their target genes by employing a wide range of molecular mechanisms [23-26].
One important theme of regulatory lncRNAs, which is supported by several experimental evidences, is that lncRNAs transcription can have profound consequences on nearby gene expression by using direct or indirect mechanisms. Transcription of a given lncRNA across the promoter region of a protein-coding gene can directly interfere with transcription factor binding to the promoter and thus prevent the expression of the neighboring gene. Such transcriptional interference mechanism has been shown to regulate key developmental decisions. One example is regulating homeotic HOX genes expression pattern and localization [27]. Even if not directly interfering with nearby promoter transcription, lncRNAs can induce histone modifications that repress transcription initiation of overlapping protein-coding gene. For example, HOTAIR (HOX Antisense Intergenic RNA) recruits the polycomb chromatin remodeling complex PRC2 inducing heterochromatin formation in specific genomic loci leading to transcriptional repression [28]. Furthermore, several lncRNAs, including the protypical lncRNA Xist (X inactive‐specific transcript), have been implicated in gene silencing or activation by guiding chromatin-modifying enzymes and associated lncRNAs to their target genes [28-29]. Alternatively, lncRNA transcription can arise in a stepwise manner from multiple sites upstream of a promoter, causing a chromatin- opening cascade, which proceed progressively toward the mRNA transcription start site acting in trans [30]. Moreover, lncRNAs can function by regulating transcription through interacting with RNA‐binding proteins [31], such as Polycomb and Trithorax group members acting as a coactivator of TFs [32]. They can also serve as precursors or as a sponge for small interfering RNAs (siRNAs) titrating their activity [33], or regulating gene expression at the post‐transcriptional editing, including the splicing level [7].
Genomic Discovery of Regulatory LncRNAs in Neonatal Heart Maturation and CHDs
Lacking sequence conservation, identification and functional characterization of lncRNAs have been challenging. However, with the combination of advanced sequencing and analytic platforms and powerful functional screening tools, these limitations can now be circumvented, leading to increased understanding of the regulatory lncRNAs roles in cardiac development and disease. Elegant studies have implicated lncRNAs in the regulation of chromatin and the control of the pluripotency network [34,35]. Other recent reports have identified Braveheart, Fender and Myheart [8,10,36,37] as critical players during heart development and disease. More recently three new players were reported in early cardiovascular development of vertebrate heart [9]. However, limited reports have focused on elucidating lncRNAs roles in neonatal heart maturation, particularly in the context of CHDs.
Aiming to improve our understanding of this important layer of genome regulation in neonatal heart maturation and their potential implication in CHDs, we and others have implemented deep RNA-sequencing and comprehensive bioinformatics analysis to explore the lncRNAs world in neonatal heart [1-4]. Neonatal lncRNAs profiling reported in our study [1] complemented two reports where deep RNA sequencing was implemented in postnatal hearts at specific time points, focusing on data from P2 and P13 hearts and from P1 and P21 hearts, respectively. Moreover, our analysis focused on lncRNA profile and correlation to mRNA in neonatal heart left and right ventricles during the critical window of perinatal circulatory transition (before and after ductal closure), which was not included in these two data sets. We also employed systems genetics tools and different methods from these two studies. Our findings provide the first delineation of transcriptome landscape in neonatal left and right ventricular chambers during three stages of fetal to neonatal transition at high level of resolution. This is the first time the neonatal heart coding genes and lncRNAs expression have been systematically analyzed in spatial-temporal manner, providing supportive evidence of the putative regulatory roles of lncRNAs in transcriptome programming, all of which may help identify functional lncRNAs of potential translational values in neonatal heart maturation in the context of CHDs.
Our genome-wide analysis pipeline highlights the dynamic nature of lncRNAs expression in neonatal heart in parallel to protein-coding genes. Hence, we learned that lncRNAs are subject to large-scale temporal variation in even a narrow, tightly regulated window, of fetal to neonatal development, suggesting a potential regulatory impact in fine-tuning overall cellular transcriptome patterns in a highly sensitive manner.
Not only do lncRNAs exhibit tight dynamic regulation, but also the tissue specificity of the lncRNAs species has surpassed that of protein-coding transcripts [3,10]. During perinatal transition, the left and right ventricle undergo significant changes in morphology and workload. To our surprise, our study revealed few lncRNAs exhibiting chamber-specific pattern. From these observations we speculate that lncRNA transcription is more intimately linked with premodial tissue and plastic cell phenotypes. Indeed, the tissue and cell specificity of lncRNAs has been shown most clearly and elegantly during cardiac progenitor differentiation and early cardiogenesis [38].
The entire dataset is disseminated through a new online resource (Neonatal Heart Maturation SuperSeries GSE85728 (http://www.ncbi. nlm.nih.gov/geo/query/acc.cgi?acc=GSE85728) for research community. In the following we summarize the lessons learned and highlighted insights that may serve to move forward regulatory lncRNAs discovery in this context.
A workflow summarizing our lncRNA discovery pipeline is shown in Figure 2. The first step in the analysis process is to assemble transcripts from the RNA-seq derived sequence reads.
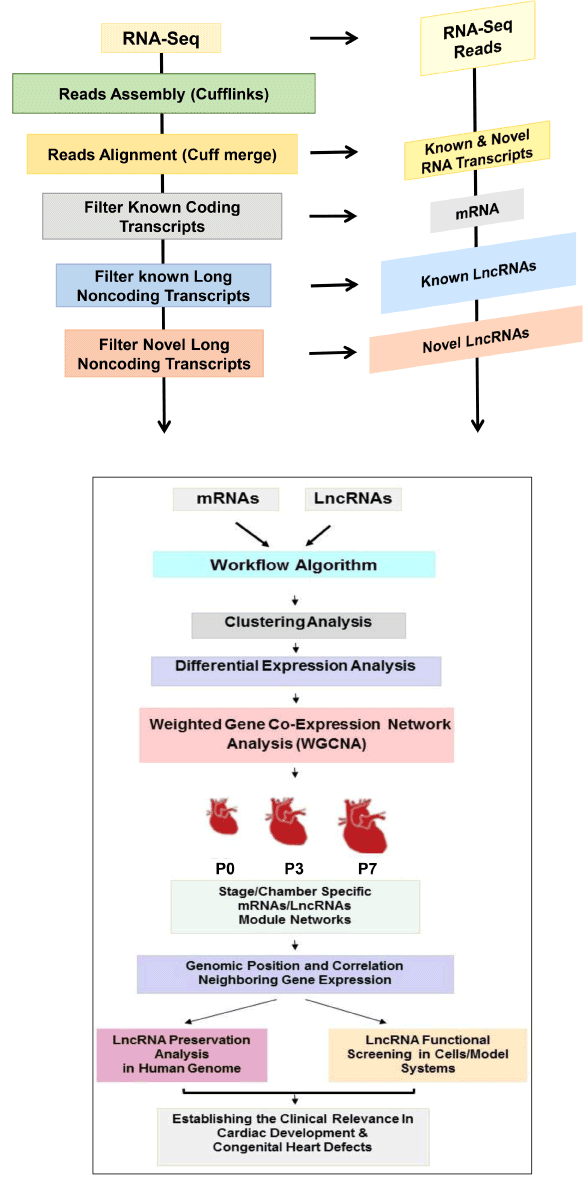
Figure 2. Regulatory LncRNA Discovery Algorithm. Schematic representation for RNA-seq data analysis pipline and systematic workflow for regulatory lncRNA discoverr in neonatal heart during perinatal circulatory transition at postnatal day 0 (P0), P3, and P7.
In our work, since we focus on identification of lncRNAs in mouse for which the genome sequences are well annotated, we have used a classical approach of first mapping RNA-Seq reads to the genome and then performing transcriptome reconstruction using the Cufflinks algorithm [20]. An alternative strategy is to perform De Novo transcript assembly directly from the sequence reads without the need of a reference genome [11]. The latter strategy is necessary when no reference genome is available. We started from three or more samples of ribosomal RNA depleted, polyA enriched mRNA to construct strand specific cDNA libraries and sequence to a depth of approximately 300 million 100x100 nt paired end reads. A minimum of three biological replicates per condition is necessary to enable any meaningful downstream statistical comparisons in a highly controlled experiment. Following transcript assembly from mapped sequence reads, the next step is to merge or align the results of the raw transcript prediction with known transcripts from a reference database of annotated transcripts such as Ensembl database [1]. This is performed using Cuff merge, which is part of the Cufflinks package.
Having identified the previously annotated transcripts, separating the transcripts into known and novel is now possible, including both protein-coding and noncoding transcripts at this stage. Following this step novel lncRNAs can be identified by filtering out those transcripts that contain an ORF exceeding 100 bp (likely protein-coding transcript, or contain a single exon (likely technical artifacts), as well as those transcripts that are below 200nt in length (as per accepted standards to define a lncRNA), and then use Gene ID to score novel transcripts for their coding potential. In our experience, using read depths of over 300 million paired-end stranded reads, we were recently able to identify more than 2800 novel lncRNAs in neonatal heart during perinatal transition stages [1]. Finally, whether a given RNA transcript exerts protein-coding potential is fundamental to the definition of lncRNA and remains challenging task. Several methods have been implemented recently to ascertain the lack of coding potentials of novel transcripts, including invitro expression and translation, codon substitution frequency analysis and ribosomal profiling among several others [39-42].
Regulatory LncRNAs in Neonatal Heart Maturation and CHDs: Functional Prediction, Challenges, and Limitation
Navigating the most difficult issue in lncRNA biology that is to depict the biological function of a putative regulatory lncRNA with poor structural conservation and very limited prior information of biochemical properties is a challenging task [17]. To overcome these difficulties, putative candidates can be prioritized for functional characterization based on a number of criteria including: a. cytoplasmic vs. nuclear expression. b. tissue-and cell type-specific expression. c. correlated expression with physiological indices. d. association with specific chromatin states. e. correlated co-expression with coding genes of functional relevance (a guilt-by-association approach). f. dynamic regulation in response to developmental or environmental stimuli. g. the existence of a human ortholog.
As previously stated, a useful clue for defining the range of interactions for a given lncRNA is determining its subcellular localization. Nuclear lncRNAs may influence transcriptional outputs through multiple mechanisms, including epigenetic modifications, interactions with transcription factors, and affecting mRNA processing or export. On the contrary, predominantly cytoplasmic lncRNAs may function through different mechanisms, including influencing the stability of an mRNA, affecting translation initiation, acting as competing endogenous RNAs, or influencing post-translational modification [23].
In our work, we utilized a systematic, stepwise, algorithm [Figure 2]. As mentioned above, many lncRNAs can remodel their local chromatin environment to regulate the expression of nearby coding genes in cis in response to developmental and environmental ques. Much of the pioneering work in transcriptome characterization during heart development was focused on differential expression studies of the lncRNAs and mRNA separately [3,4]. In our work, we furthered our analysis by performing parallel, unsupervised weighted gene co- expression network analysis (WGCNA) [43,44] along the 3 time points on the protein-coding and long noncoding transcript (a guilt-by-association approach). Hence, our integrative, systems-based, approach revealed several lncRNA and mRNA co-expression modules that are stage-specific and coordinately shared in both ventricles. These modules included 33 members (11 known and 22 novel) of abundantly expressed lncRNAs, which exhibited concordant regulation with their corresponding mRNA modules in a stage-specific fashion reflecting the rapid adaptation of cardiovascular system to sharp changes in circulation and postnatal environment.
After constructing lncRNA/mRNA modules, we examined lncRNAs correlation with neighboring gene expression. In total, we reported 114 lncRNAs, including 12 novel lncRNAs that showed significantly correlated expression pattern with a neighboring mRNA. Next, we identified their potential orthologues in human. Then, we validated the expression of candidate lncRNA/mRNA pairs by quantitative RT-PCR in neonatal mouse heart and human CHDs samples to examine expression levels and tissue specificity. Finally, we verified that the regulatory relationship of the top four correlated lncRNA/mRNA pairs (Ucp2-lncRNA, n420212, FUS- lncRNA, and Ppp1r1b-lncRNA and their partner mRNAs, UCP3, KCNB1, TRIM72, and TCAP, respectively) is preserved in human CHDs samples. Furthermore, the expression ratio of Ppp1r1b- lncRNA/Tcap segregated CHDs based on their structural phenotypes [1].
To better understand the physiological roles of regulatory lncRNAs in neonatal heart development, we employed antisense oligos (GapmeR) to achieve targeted Ppp1r1b-lncRNA silencing in C2C12 cells, a muscle myoblast cell line. Our findings demonstrated that the Ppp1r1b-lncRNA modulates its neighboring partner gene Tcap (titin cap), which is important for sarcomeric integrity and function and other muscle regulatory factors, leading to promoting myogenic differentiation process.
Our findings hinted at a higher order regulatory architecture for controlling gene expression at the genome-wide and the transcript-specific levels, in which the lncRNAs were demonstrated to have potential regulatory roles in neonatal heart maturation and CHDs. These insights provide essential foundations that would advance our current understanding of the gene regulatory network and pave the way to investigate the underlying mechanisms. Having demonstrated tightly conserved regulation in human CHDs, we believe that some of these lncRNAs play important intrinsic regulatory function. Further experimental studies to characterize their diverse mechanisms may provide a physiological basis for future investigations in pathological maturation and CHDs. At the mechanistic and technical levels, in addition to using RNA interference and modified antisense oligonucleotide approaches, new approaches for targeting lncRNAS are needed to overcome several challenges. For example, large fraction of newly discovered lncRNAs in heart act as regulators of the epigenome and nuclear architecture in cis at their site of production. In particular, the regulatory function of lncRNAs derived from active enhancers act primarily at their endogenous site of production. Therefore, modulation of the nascent transcript is critical. Furthermore, identifying chromatin marks and histone modifications at gene promoters and further dissecting the molecular mechanisms requires employing sophisticated molecular genetics tools, such as chromatin isolation by RNA purification (ChIRP) [45] and capture hybridization analysis of RNA targets (CHART) [46]. Moreover, genome-editing technologies can serve to modify cardiovascular lncRNA expression in vivo. The lncRNA field will certainly benefit from new techniques to generate targeted mutations in the genome, such as Cas9/CRISPR systems. Finally, hiPSCs (human induced pluripotent stem cells) can provide powerful platforms for future studies designed to establish the mechanisms of human cardiac lncRNA and their potential regulators in development and disease, and then to explore their potential applications as putative disease modifiers, diagnostic biomarkers, and therapeutic targets in pathological maturation of neonatal heart and CHDs.
This work was supported by grants from the American Heart Association Career Development Award 18CDA34110414; NIH/NHLBI Bridge Fund 1R56HL146738-01; the Department of Defense-Congressionally Directed Medical Research Programs W81XWH-18-1-0164; the UCLA- Children’s Discovery Institute and Today and Tomorrow Children’s Fund; and the David Geffen School of Medicine Cardiovascular Theme Research Innovation Seed Grant to M. Touma.
We refer interested researchers to gene expression data sets deposited within the Gene Expression Omnibus repository (www.ncbi.nlm.nih.gov/geo) under Neonatal Heart Maturation SuperSeries GSE85728 (http://www.ncbi. nlm.nih.gov/geo/query/acc.cgi?acc=GSE85728). All unique materials, resources and reagents are available on request by qualified researchers for their own use.
The author declares no conflict of interest.
- Touma M, Kang X, Zhao Y, Cass AA, Gao F, et al. (2016) Decoding the long noncoding RNA during cardiac maturation: a roadmap for functional discovery. Circ Cardiovasc Genet 9: 395-407. [Crossref]
- Lee JH, Chen Gao, Peng G, GreerC, Ren R, et al. (2011) Analysis of Transcriptome Complexity through RNA Sequencing in Normal and Failing Murine Hearts. Circulation research 109: 1332-1334. [Crossref]
- Gan J, Sonntag HJ, Tang MK, Cai D, Lee KK (2015) Integrative analysis of the developing postnatal mouse heart transcriptome. PLoS One 10: e0133288. [Crossref]
- Matkovich SJ, Edwards JR, Grossenheider TC, de Guzman Strong C, Dorn GW 2nd (2014) Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc Natl Acad Sci USA 111: 12264-12269. [Crossref]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, et al. (1992) The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515-526. [Crossref]
- Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, et al. (2014) Defining functional DNA elements in the human genome. Proc Natl Acad Sci USA 111: 6131-6138. [Crossref]
- Wang Z, Wang Y (2015) Dawn of the Epi-LncRNAs: new path from Myheart. Circ Res 116: 235-236. [Crossref]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, et al. (2013) Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152: 570-583. [Crossref]
- Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, (2015) Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 131: 1278-1290. [Crossref]
- Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, et al. (2013). The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24: 206-214. [Crossref]
- Friedrichs F, Zugck C, Rauch GJ, Ivandic B, Weichenhan D, et al (2009) HBEGF, SRA1, and IK: three cosegregating genes as determinants of cardiomyopathy. Genome Res 19: 395-403. [Crossref]
- Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, et al. (2006) Identification of a novel non- coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51: 1087-1099. [Crossref]
- Olson EN, Srivastava D (1996) Molecular pathways controlling heart development. Science 272: 671-676. [Crossref]
- Han X, Zhang J, Liu Y, Fan X, Ai S, et al. (2019). The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development 146: dev176198. [Crossref]
- Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, et al. (2016) Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539: 433-436. [Crossref]
- Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, et al. (2014) Defining functional DNA elements in the human genome. Proc Natl Acad Sci USA 111: 6131-6138. [Crossref]
- Wang J, Zhang J, Zheng H, Li J, Liu D, et al. (2004) Mouse transcriptome: neutral evolution of 'noncoding' complementary DNAs. Nature 431: 7010. [Crossref]
- Derrien T, Vaysse A, Andre C, Hitte C (2012) Annotation of the domestic dog genome sequence: finding the missing genes. Mamm Genome 23: 124-131. [Crossref]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, et al. (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915-1927. [Crossref]
- Ritter O, Haase H, Schulte HD, Lange PE, Morano I (1999) Remodeling of the hypertrophied human myocardium by cardiac bHLH transcription factors. J Cell Biochem 74: 551-561. [Crossref]
- Guttman M, Amit I, Garber M, French C, Lin MF, et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223-227. [Crossref]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, et al. (2006) Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 16: 11-19. [Crossref]
- Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904-914. [Crossref]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA 105: 716-721. [Crossref]
- Mercer TR, Dinger ME, Mattick JS (2005) Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155-159. [Crossref]
- Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, et al. (2009) Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol 15; 182. [Crossref]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311-1323. [Crossref]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750-756. [Crossref]
- Ogawa Y, Sun BK, Lee JT (2008) Intersection of the RNA interference and X-inactivation pathways. Science 320: 1336-1341. [Crossref]
- Mercer TR, Mattick JS (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 20: 300-307. [Crossref]
- Wang X, Arai S, Song X, Reichart D, Du K et al. (2008) Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454: 126-130. [Crossref]
- Feng J, Bi C, Clark BS, Mady R, Shah P, et al. (2006) The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 20: 1470-1484. [Crossref]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A (2007) Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445: 666-670. [Crossref]
- Paralkar VR, Mishra T, Luan J, Yao Y, Kossenkov AV, et al. (2014). Lineage and species-specific long noncoding RNAs during erythro- megakaryocytic development. Blood 123: 1927-1937. [Crossref]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, et al. (2010) Simple combinations of lineage-determining transcription factors prime cis- regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576-589. [Crossref]
- Yan L, Tang M, Guo H, Yang L, Wu J, et al. (2013) Single-cell RNA seq profiling of human preimplantation embryos and embryonic stem cells. Nature Structural and Molecular Biology 20: 9. [Crossref]
- Han P, Li W, Lin CH, Yang J, Shang C, et al. (2014) A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514: 102-106. [Crossref]
- Rizki G, Boyer LA (2015) Lncing epigenetic control of transcription to cardiovascular development and disease. Circ Res 117: 192-206. [Crossref]
- Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, et al. (2007) Revisiting the protein- coding gene catalog of Drosophila Melanogaster using12 fly genomes. Genome Res 17: 1823-1836. [Crossref]
- Lin MF, Deoras AN, Rasmussen MD, Kellis M (2008) Performance and scalability of discriminative metrics for comparative gene identification in 12 Drosophila genomes. PL0S Comput Biol 4: e1000067. [Crossref]
- Pueyo JI, Couso JP (2011) Tarsal-less peptodes control Notch signaling through the Shavenbaby transcription factor. Dev Biol 355: 183-193. [Crossref]
- Ingolia NT, Lareau LF, Weisman JS (2011) Ribosome profiling of mouse embryonic stem cells reveal the complexity and dynamics of mammalian proteomes. Cell 147: 789-802. [Crossref]
- Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Gent Mol Biol 4: 17. [Crossref]
- Langfelder P, Zang B, Horvath S (2008) Defining clustering from a hieratical cluster tree: The dynamic tree cut package for R. Bioinformatics 24: 719-720. [Crossref]
- Chu C, Quinn J, Chang HY (2012) Chromatin isolation by RNA purification (ChIRP) J Vis Exp 61: 3912. [Crossref]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44: 667-678. [Crossref]


