Aim: Colorectal cancer is a potentially curable malignancy; but it remains as a fatal disease since it’s the second most common cause of cancer-related death globally. Currently, 5-Fluorouracil is the main chemotherapeutic agent used in combined therapies for the adjuvant and palliative treatment of colorectal cancer. Genetic polymorphisms have important roles in individual differences which occur in response to cancer chemotherapy and toxicity. The aim of this study is to evaluate association of 5-Fluorouracil metabolism-related MTHFR-677C>T, MTHFR-1298A>C, GSTP1-313A>G and TSER single nucleotide polymorphisms with chemotherapy response and toxicity.
Methods: One hundred chemotherapy naive colorectal cancer patients were enrolled and their biochemical parameters evaluated. Mutation analysis of MTHFR-677C>T, MTHFR 1298A>C, TSER and GSTP1-313A>G genes were studied in Molecular oncology laboratory of Medical Biology Department.
Results: There was an association between GSTP1-313A>G A/G genotype and smoking. MTHFR-677C>T T/T ve C/T genotypes were related to neutropenia, high levels of CEA and CRP. MTHFR-1298A> C C/C genotype was associated with high levels of CRP and progression. High levels of CEA, CRP, diarrhea-constipation and neutrophilia were related to TSER-3R/3R and 2R/3R genotypes. In survival analysis, shorter disease free survival was observed for MTHFR-1298A>C single nucleotide polymorphism C/C genotype in early stage patients.
Conclusion: In this study, we have investigated single nucleotide polymorphisms of specific enzymes involving in activity and metabolism of 5-FU and their association with response to chemotherapy and toxicity. The limited number of patients has led us to the conclusion that our results should be supported by more comprehensive multi-centered studies.
colorectal cancer, GSTP, MTHFR, TSER, single nucleotide polymorphism, 5-Fluorouracil, toxicity, chemotherapy response
Colorectal cancer is a potentially curable malignancy; but it still remains as a fatal disease since it’s the second most common cause of cancer-related death globally with serious damage to human health [1]. Gene polymorphisms involve in both pathogenesis of colorectal cancer and metabolism of the drugs used in treatment (Fluorouracil (5-FU), oxaliplatin and irinotecan). While certain polymorphisms may lead to changes in effectiveness of the drugs, some others substantially increase toxicities of these drugs [2]. Thus, identification of polymorphisms may be effective in predicting clinical response to chemotherapy and chemotherapy-related toxicities in several types of cancer [3,4].
5-Fluorouracil acts via fluorodeoxyuridine monophosphate (FdUMP). This molecule inhibits thymidylate synthase (TS) [5]. Reduction in methylenetetrahydrofolate reductase (MTHFR) enzyme activity may lead to an increase in intracellular 5,10-methylenetetrahydrofolate (5,10-MTHF) level and thus may increase FU cytotoxicity. MTHFR gene is localized to chromosome 1p36.3 [6]. Two types of single-nucleotide polymorphisms (SNPs) frequently develops on this enzyme: C677T (rs1801133) and A1298C (rs1801131) [7]. Alanine and valine exchanges their position as a result of C677T transition and thus form a more thermo-labile protein which reduces enzyme activity [8]. A1298C variant (Glu429Ala) also may reduce MTHFR enzymatic activity by leading to a missense mutation [9]. In C677T homozygous (TT) or heterozygous (CT) genotypic conditions MTHFR enzyme activity is downregulated and blood homocysteine level increases [10]. When A1298C enzyme changes to homozygous (CC) or heterozygous (AC) form from its normal (AA) form a reduction in MTHFR activity is observed but thermo-labile protein is not formed [11]. Reduced enzyme activity results with high levels of 5,10-MTHF and thymidine and thus leads to an increase in DNA synthesis and repair. Thus, MTHFR polymorphisms are considered as protective against tumorigenesis [12]. MTHFR SNPs and effectiveness of 5-FU therapy have been assessed in experimental and clinical studies [13,14].
Chemical carcinogens such as polycyclic aromatic hydrocarbons and heterocyclic aromatic amines have been found to be associated with colon cancer [15]. These compounds are found in tobacco smoke or meat products cooked in high heat and open blaze [16]. As a result of metabolic activation of these compounds highly reactive mutagens occur and interact with DNA bases [15]. These compounds may transform to stable products after detoxification by glutathione S transferase (GSTP1) and uridine diphosphate-glucuronosyltransferase (UDPGT) [17]. As a result of transformation of adenine (A) to guanine (G) at nucleotide 313 in exon 5 of GSTP1 gene a change in amino acid 104 as substitution of isoleucine by valine occurs. This change at a site near to hydrophobic binding region of the protein decreases affinity for electrophilic compounds [18]. Various studies on drug metabolism have shown that GSTP1 SNP may be used in predicting clinical outcome in patients receiving 5-FU based chemotherapy for treatment of metastatic colorectal cancer [19,20].
Functional polymorphisms in thymidylate synthase gene may increase risk of various types of cancer by exerting an impact on nucleotide synthesis by changing effectiveness of the enzyme [21]. In thymidylate synthase promoter end region (TSER) dual (2R) and triple (3R) recurrent polymorphisms are described [22]. In vitro and in vivo studies have reported that TS expression is TSER genotype dependent and its 3R allele lead to TS overexpression [22]. Thus, various epidemiological studies related with colorectal cancer risk and TSER 2R/3R polymorphism that has an impact on folate metabolism have been conducted, however the results are inconsistent [23,24].
MTHFR, TS and GSTP1 enzymes that involve in 5-Fluorouracil metabolism determine drug effectiveness and toxicity. MTHFR 677C>T transition and 1298A>C transversion mutations are shown to be associated with high cytotoxic activity of fluorouracil in colon and breast cancer cells [25]. It’s widely accepted that increased TS expression is the main molecular mechanism that is responsible from 5-FU resistance and its use as a potential prognostic and predictive marker is suggested [26].
The aim of this study is to evaluate association of chemotherapeutic agent 5- FU metabolism-related TSER, MTHFR- 677C>T, MTHFR 1298A> C and GSTP1-313A>G SNPs with chemotherapy response and toxicity.
Patients
Medical Ethics committee of Pamukkale University Medical Faculty has approved our study in April 22, 2014. In this study 100 patients who have been treated in Medical Oncology Department of our hospital between April 04, 2014 and October 22, 2014 with pathological diagnosis of colon and rectum cancer were evaluated. Patients with WHO performance status 0, 1, 2 at all stages (stage I, II, III and IV) were included into the study. Patients 20-80 years old who have given written informed consent were included. Performance status 3 or worse with brain metastasis or suspected of brain metastasis whose age is 80 years or older or 20 years or younger and who haven’t signed informed consent form were excluded from the study.
Laboratory tests
Blood samples were obtained from patients between 08:00-09:00 A.M. after at least 8-12 hours of overnight fasting before treating patients. From the blood samples hemogram results were obtained by CELL-DYN 3700 Systems and CELL-DYN Sapphire instrument and albumin, C-reactive protein, CRP, lactate dehydrogenase (LDH), carcinoembryonic antigen (CEA), CA 19.9, albumin results were obtained by Roche/Hitachi Cobas c Systems, Cobas c 501 and Roche/Hitachi Cobas c Systems, e 601 Module instruments.
DNA isolation
Blood samples of 100 patients under assessment were obtained to CBC tubes. These patients referred to Medical Oncology Department for having treatment because of pathological diagnosis of colon and rectum cancer and 200 ul blood was obtained and blood samples were used for DNA isolation according to QIAGEN EZ1 DNA blood 200 ul kit protocol by using EZ1 Advanced XL instrument.
Mutation Analysis
Mutation analysis of MTHFR- 677C> T, MTHFR 1298A> C, TSER and GSTP1-313A>G genes were studied in Molecular Oncology Laboratory of Medical Biology Department.
The extracted DNAs were sequenced by pyrosequencing method which is a sequence method gives result effectively with as less as 15 ng DNA. This method is based on real time non-electrophoretic system. PCR reaction composed of 20 pmol of each primer, 0.2 mmol each of deoxy-nucleotide triphosphates, 1.5 mmol/L MgCl2, and 1.25 U of FastStart Taq DNA polymerase (Roche, Basel, Switzerland) as total final volume of 50 μL. PCR condition consisted of an initial denaturation step at 95°C for 15 minutes, followed by 42 cycles at 95°C for 20 seconds, 54°C for 30 seconds, 72°C for 20 seconds, and a final extension step at 72°C for 5 minutes. The PCR products (10 μL) were analysed by electrophoresis in a 3% agarose gel to confirm successful amplification.
After validation of the PCR products on the gel, 40 μL of the products were bound to streptavidin Sepharose HP (GE Healthcare, Waukesha, Wisconsin), purified, washed, and denatured using a 0.2 mol/L sodium hydroxide solution, and washed again. The pyrosequencing was performed on a PyroMark ID system (Qiagen) according to the manufacturer’s instructions and analyzed in AQ mode of the PyroMark software.
Single nucleotide polymorphism analysis was done by using pyrosequencing method. Pyrosequencing method is mainly based on identifying pyrophospate (PPi) that is liberated during DNA synthesis. Fluorescein-based sequencing is used. This method is a non-electrophoretic fluorescein system. The rationale of the system is basically as follows: during synthesis of sequencing primers on DNA template strand inorganic pyrophosphates are released and during conversion of ADP to ATP by ATP sulfurase enzyme luciferin is converted to oxyluciferin and that generates a visible light. When inorganic pyrophosphates are exposed to this light the light can be seen as peaks in the diagram. The distinctive criterion here is addition of adenine, guanine, thymine and cytosine nucleotides to the medium on at a time. In the Q24 sequencing device whichever of the nucleotides reacts and light occurs the resultant peak in the system belongs to that nucleotide. A substrate is added to the medium to accelerate the system. For calibration of the system the measurement of enzyme and substrate compound is given in the beginning. This result provides information about whether the system is running or not. Also, nucleotide triphosphates are degraged to mono or dinucleotidephospates (dNTP) and ATP to mono or diphosphate by apyrase enzyme in order to prevent false peak in the medium after the reaction. When new dTNP is given once more, then the reaction starts again.
PCR was performed by qiagen kit to understand whether TSER gene tandem repeat number is double or triple. PCR protocol; 1 cycle of denaturation for 15 minutes at 95°C, annealing 20 secs 95°C-30 sec 53°C -20 sec 72°C 42 cycle and last extension 72°C 5 minutes. PCR products were visualized on agarose gel.
Statistical analysis
Statistical analysis is performed by using Statistical Package for Social Sciences version 16.0 (SPSS- 16.0, for windows) package program. The results were assessed within 95% confidence interval. P<0.05 was considered statistically significant. For comparing genetic mutations with clinic-pathological characteristics of patients χ2 and Mann Whitney-U test were used.
Clinic and pathological characteristics such as sex, age, tobacco use, diagnosis, stage, CEA, CA 19-9, LDH, CRP levels, leukocytosis, neutrophilia, thrombocytosis, anemia, thrombocytopenia, neutropenia, mucositis, diarrhea-constipation, nausea-vomiting, neurotoxicity and hand-foot syndrome were used as co-variates for the comparison. Spearman and Pearson correlation tests were used for correlation analysis. For overall survival (OS) and progression free survival (PFS) and time-survival curves Kaplan Meier method was used. For analysis of factors that have an impact on survival and progression logistic regression was used.
Our study was conducted on 100 patients with early, advanced and metastatic stage colorectal cancer diagnosis who were treated with 5-FU-Leucoverin (LV), 5-FU-LV-oxaliplatin (FOLFOX) and 5-FU-LV-irinotecan (FOLFIRI) based combination chemotherapy regimens. Sixtyfour patients were male (64%) and 36 females (36%). Mean age of the patients was 61.81±10.97 years and median age was 63 years [21-28]. Main clinical, genetic and demographic characteristics of patients were shown in Tables 1 and 2. Statistically significant results between SNPs and clinical characteristics were shown in Table 3.
Table 1. Clinical and demographic characteristics of the patients
Characteristics
No (%) |
Early stage
(n=77) |
Metastatic stage
(n=23) |
Overall group
(n=100) |
Age (year) |
62.8±10.9 |
58.4± 10.4 |
61.81±10.97 |
Sex
Female
Male |
28 (36.4)
49 (63.6) |
8 (34.8)
15 (65.2) |
36 (36)
64 (64) |
Diagnosis
-Colon
-Rectum |
55 (71.4)
22 (28.6) |
17 (73.9)
6 (26.1) |
72 (72)
28 (28) |
Performance status
ECOG (0-1-2) |
77 (77) |
23 (23) |
100 (100) |
Tobacco smoking
-Smoker
-Ex-Smoker |
38 (49.4)
39 (50.6) |
13 (56.5)
10 (43.5) |
51 (51)
49 (49) |
Table 2. Gene polymorphism analysis results of the patients
Single nucleotide polymorphism No (%) |
Early stage
(n=77) |
Metastatic Stage
(n=23) |
Overall group
(n=100) |
 MTHFR -677C>T MTHFR -677C>T
- Mutant
- Heterozygous
- Wild type |
8 (10.4)
29 (37.7)
40 (51.9) |
2 (8.7)
10 (43.5)
11 (47,8) |
10 (10)
38 (38)
52 (52) |
MTHFR- 1298A>C
-Mutant
-Heterozygous
-Wild type |
15 (19.5)
24 (31.2)
38 (49.4) |
3 (13)
8 (34.8)
12(52.2) |
18 (18)
30 (30)
52 (52) |
TSER
-Mutant (3R/3R)
-Heterozygous (2R/3R)
-Wild type (2R/2R) |
22 (28.6)
29 (37.7)
26 (33.8) |
8 (34.8)
10 (43.5)
5 (21.7) |
31 (31)
37 (37)
32 (32) |
GSTP1-313A>G
-Mutant
-Heterozygous
-Wild type |
3 (3.9)
33 (42.9)
41 (53.2) |
2 (8.7)
10 (43.5)
11 (47.8) |
4 (4)
43 (43)
53 (53) |
Table 3. Statistically significant results between gene polymorphisms and clinical characteristics
Polymorphism |
Clinical features |
p value |
MTHFR-677C> T |
Tumor location |
Rectum |
Colon |
|
All patients |
TT %60 |
%40 |
0.043 |
Early stage |
TT %75 |
%25 |
0.003 |
CRP levels |
High |
Low |
|
Early stage |
TT %100 |
--- |
0.016 |
CEA levels |
High |
Low |
|
Metastatic |
TT + CT %100 |
--- |
0.03 |
|
Yes |
No |
|
Metastatic |
TT + CT %70 |
%30 |
0.021 |
MTHFR-1298A> C |
CRP levels |
High |
Low |
|
Early stage |
CC %86.7 |
%13.3 |
0.029 |
|
|
|
|
Progression |
Yes |
No |
|
Early stage |
CC %66.7 |
%33.3 |
0.012 |
GSTP1-313A>G |
Smoking |
Yes |
No |
|
Metastatic |
AG %61.5 |
%38.5 |
0.03 |
TSER 2R / 3R |
CRP levels |
High |
Low |
|
All patients |
3R + 2R-3R %78.4 |
%11.6 |
0.018 |
CEA levels |
High |
Low |
|
All patients |
3R + 2R-3R %75 |
%25 |
0.006 |
Diarrhea–constipation |
Yes |
No |
|
Early stage |
3R %80 |
%20 |
0.024 |
Hypoalbuminia |
Yes |
No |
|
Early stage |
3R + 2R-3R %86.7 |
%13.3 |
0.047 |
Neutropenia |
Yes |
No |
|
Early stage |
3R + 2R-3R %90.9 |
%9.1 |
0.04 |
Survival analysis
In 42 of (42%) 100 patients progression has occurred. In early stage patients mean progression-free survival (PFS) was 30.6 months and mean overall survival (OS) was 39.3 months. In metastatic patients mean PFS was 14 months and mean OS was 23 months.
Progression-free survival analysis according to MTHFR 1298A>C SNP has revealed that PFS rate and mean PFS was 33.3% and 36 months in mutant patients and 76.3% and 67 months in patients with wild genotype and 66.7% and 114 months in heterozygous group respectively. The difference between subgroups was statistically significant (Log-Rank= 0.013) (Figure 1).
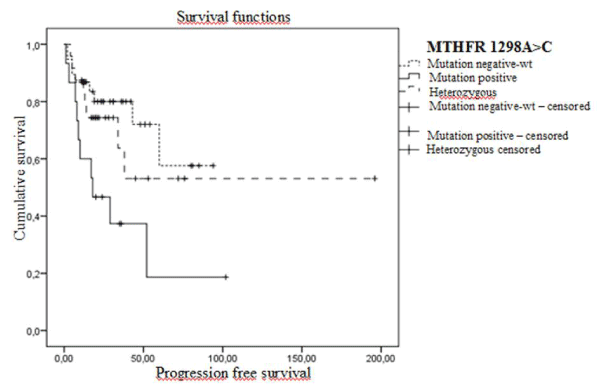
Figure 1. Progression-free Kaplan Meier survival curves in early stage patients according to MTHFR 1298A>C polymorphisms (wt: A/A genotype, heterozygous: A/C genotype and mutant: C/C genotype).
Progression-free survival analysis of MTHFR 1298A>C SNP patients groups has revealed statistically significant differences. For mutant patients disease-free survival rate and mean PFS was 33.3% and 36 months and in wild and heterozygous patients 72.6% and 123 months respectively (Log-Rank=0.004). Disease-free survival curves are shown in Figure 2.
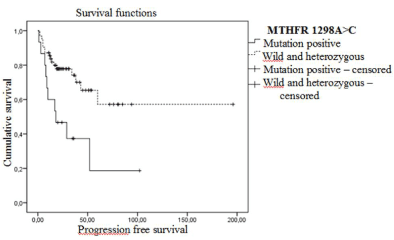
Figure 2. Progression-free Kaplan Meier survival curves in early stage patients according to MTHFR 1298A>C polymorphism (wt: A/A genotype, heterozygous: A/C genotype and mutant: C/C genotype).
In the survival analysis of TSER, MTHFR- 677C>T SNP patient groups mean OS was shorter in mutant patients relative to wt and heterozygous patients but the difference wasn’t statistically significant.
Analysis of MTHFR 1298A> C SNP in metastatic patients has shown that mutant patients had shorter mean PFS relative to wt and heterozygous patients but the difference wasn’t statistically significant. In the survival analysis of TSER, MTHFR- 677C>T and GSTP1-313A polymorphism subgroups OS rate and mean OS wasn’t statistically different.
Colorectal cancer, is the second most common cause of cancer-related death just after lung cancer. Despite recent advances in colorectal cancer therapy and our increasing understanding about the molecular pathways, our knowledge is still insufficient to answer the question of why some patients respond to treatment but some others not. At this point, metabolisms of the drugs used for chemotherapy and resistance mechanisms related with these drugs become important. Pharmacogenetic studies related with cancer drugs have shown that there is association between specific genetic variants of enzymes involve in drug metabolism and drug response and adverse effects. Thus, further information is needed about specific genetic variants of enzymes involve in drug metabolism and genetic abnormalities that has impact on response to treatment.
Alterations in MTHFR enzyme activity lead to changes in effectiveness of 5-FU [26,28]. Multiple polymorphisms are described in MTHFR gene. MTHFR- 677C>T and MTHFR 1298A> C single nucleotide polymorphisms are the two important polymorphisms that are most associated with enzyme activity alterations. Zhao’s meta-analysis has reported that in the presence of MTHFR- 677C>T mutation, 14% reduction in colorectal cancer risk was observed [7]. In our study MTHFR- 677C>T mutant patients’ ratio was found as 10% which is in line with literature.
In our study, significant associations were observed with MTHFR-677C>T polymorphism mutant genotype. In the early stage patient group high CRP levels and in the metastatic patient group high CEA levels were associated with mutant genotype. We couldn’t observe any study showing association of these clinical parameters and MTHFR- 677C>T SNP in the English literature. Ours is the first study in this area.
Association between MTHFR- 677C>T SNP and toxicity was examined in our study; neutropenia was observed in mutant and heterozygous metastatic patients and the relation was statistically significant. Lee, et al. have shown MTHFR- 677C>T SNP mutant genotype is associated with high risk of neutropenia on 292 colorectal cancer patients [29]. Yeh, et al. have also identified significant correlation between MTHFR- 677C>T SNP and neutropenia [30]. In another study by Chua, et al. MTHFR- 677C>T SNP was found to be associated with increased risk of diarrhea [31]. MTHFR enzyme activity decreases in the presence of MTHFR- 677C>T mutant genotype and consequently 5-FU activity and toxicity increases. Thus, patients with MTHFR mutant genotype manifest more toxicity symptoms such as neutropenia. In colorectal cancer patients, it’s clear that identifying MTHFR- 677C>T SNP may contribute substantially to optimization and individualization of chemotherapy. Several studies have suggested that SNP may be related with progression, reduced response to chemotherapy and shorter OS [32]. Given an increase in CEA and CRP levels indicate presence of progression, it may be expected that in mutant patients and higher CEA and CRP levels reduced chemotherapy response and shorter OS may be observed. However, there are some other studies that couldn’t find any association between this SNP and progression or shorter OS, on the contrary there are studies even reporting increased response to chemotherapy with presence of MTHFR- 677 T/T SNP [14,32]. Under the light of all these information from the literature it may be concluded that prospective, larger randomized trials investigating MTHFR- 677 C>T SNP are needed.
MTHFR 1298A> C SNP is one of the most important polymorphism related with alteration in enzyme activity. Zhao’s meta-analysis has reported 18% colorectal cancer reduction risk in the presence of MTHFR- 1298A>C mutation [7]. In our study mutant group ratio was also 18% which is in line with the literature.
In our study, MTHFR 1298A>C SNP mutant genotype was found to be significantly related with disease progression and higher CRP levels. Etienne, et al. have found that MTHFR 1298A>C mutant genotype is associated with shorter OS and colorectal cancer patients with this genotype (11 out of 12) didn’t respond to treatment [14]. In a study conducted by Chang, et al. patients with MTHFR 1298 SNP wild genotype OS was found to be longer [33]. In some other studies no correlation was found between MTHFR 1298A>C SNP and response to chemotherapy or survival [34,35]. Association of higher CRP levels and systemic inflammation with cancer development and cancer progression was investigated in substantial number of studies. Helzlsouer, et al. have shown that higher CRP levels and systemic inflammation are risk factors for development of colon and prostate cancer [36]. Studies on patients with esophageal cancer have shown presence of association between higher CRP levels and progression [37-39]. Intervention on inflammatory process and inflammatory pathways may reduce risk of cancer development and progression. To identify patients with MTHFR 1298A>C SNP mutant genotype and higher CRP levels may help predicting progression and may prompt use of anti-inflammatory therapies for preventing progression.
2021 Copyright OAT. All rights reserv
Early stage patients with MTHFR 1298A>C SNP mutant genotype was found to be associated with shorter PFS in our study. Yeh, et al. reported significantly shorter OS with mutant genotype rectal cancer patients [30]. Afzal, et al. have conducted a study in 331 patients having adjuvant 5-FU chemotherapy and couldn’t find any association between MTHFR 1298A>C genotypes and DFS [40]. Capitain, et al. also found no association between MTHFR 1298A>C genotypes and DFS [41]. In a study by Zhang, et al. patients with MTHFR 1298 mutant genotype had a shorter DFS but statistical difference couldn’t be reached [33]. On the other hand, Etienne, et al. stated mutant genotype was associated with shorter OS [14]. It’s known that functional differences of MTHFR variants are related with plasma folate levels. The differences between results may be due to the difference in folate levels of the patients and consequent functional differences between MTHFR variants.
Currently, 5-FU is a chemotherapeutic agent used both in adjuvant and palliative treatment of colorectal cancer. Thymidylate synthase enzyme is vital for DNA replication and it’s considered as the primary intracellular target for fluoropyrimidines. Level of TS within the tumor tissue is important, because in patients with colorectal cancer treated with 5-FU based chemotherapy, lower TSER expression level within the tumor tissue is correlated with response to chemotherapy and also higher toxicity and longer survival. TSER promoter possesses 28-bp tandem, repeated sequence and it’s often observed as 2R-double repeat or 3R-triple repeat alleles. In the studies comparing 3R/3R cells with 2R/2R cells, it has been shown that in 3R/3R cells’ TSER mRNA is much more overexpressed. As a result of TS increase FU-metabolism increases and this in turn reduces effectiveness of the drug and alleviates adverse effect profile [42]. In our study, there was significant association in patients between TSER mutant and heterozygote genotypes and higher CRP and CEA levels; lower albumin levels in early stage patients; neutrophilia and higher CRP levels and in the metastatic patient group. However, there is no study in the literature supporting these results. On the contrary of other studies, in our study 80% of patients with 3R/3R genotype had diarrhea (p=0.024). In a study by Lecomte, et al. on 90 patients with a diagnosis of colorectal cancer, in patients with mutant genotype toxicity rate was 4% but in wild genotype toxicity rate was 43% and the most common toxicity was diarrhea [42]. Schwab, et al. conducted a study over 620 patients, mutant and heterozygote genotype was also found to be associated with less severe toxicity and less diarrhea [43]. There is substantial number of large studies regarding TSER and conflicting results were obtained in most of these studies [44-46]. Number of patients, differences in laboratory methods and treatment regimens may have an impact on these results. Still, predictive role of TSER in 5-FU toxicity should be investigated in larger population studies.
GSTP1 is a member of glutathione S-transferase family. It prevents DNA damage and formation of DNA insertion by conjugating inactive xenobiotics with glutathione and has a role in detoxification pathways of various drugs. Structural polymorphisms of GSTP1 lead to reduction in enzyme activity and thus decrease in detoxification activity and results with DNA damage [47]. In our study, 61.5% of tobacco smokers in the metastatic patient group were belonging to GSTP1- 313A>G SNP heterozygous genotype. Soyaa, et al. have conducted a study in 408 respiratory tract cancer patients and have found a strong interaction between tobacco smoking and tobacco chewing patients and GSTP1 mutant genotype; and determined that this interaction is associated with an increase in cancer risk [48]. Tobacco smoking, alcohol use and exposure to occupational toxins and carcinogens are known to cause 90% of all cancer types including colorectal cancer [49]. Exposure to these factors in addition to genetic factors plays a major role in etiology of colorectal cancer. Individual differences in sensitivity to any environmental exposure may be genetically based. The variation in GSTP1 gene may partially explain host sensitivity in metabolic activation and detoxification of an environmental carcinogen such as cigarette.
In this study, we have investigated TSER, MTHFR- 677C>T, MTHFR 1298A> C and GSTP1-313A>G SNPs of specific enzymes involving in activity and metabolism of 5-FU which is the primary drug used in treatment of colorectal cancer patients and their association with response to chemotherapy and toxicity. The limited number of patients has led us to the conclusion that our results should be supported by more comprehensive multi-centered studies.
- Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, et al. (2017) Global patterns and trends in colorectal cancer incidence and mortality. Gut 66: 683-691. [Crossref]
- Chan AT, Giovannucci EL (2010) Primary prevention of colorectal cancer. Gastroenterology 138: 2029-2043. [Crossref]
- Libutti SK, Saltz LB, Tepper JE (2008) Section 12: Colon cancer. In: DeVita VT, Lawrence TS, Rosenberg SA. Devita, Hellman & Rosenberg's Cancer: Principles & Practice of Oncology. (8th ed.) Philadelphia, Lippincott Williams & Wilkins, pp: 1232–1285.
- Libutti SK, Saltz LB, Tepper JE (2011) Chapter 89: Cancer of the Colon. DeVita, Hellman, and Rosenberg's Cancer: Principles & Practice of Oncology. (9th ed.) Philadelphia, Lippincott Williams & Wilkins, pp: 1084-1153.
- Chéradame S, Etienne MC, Chazal M, Guillot T, Fischel JL, et al. (1997) Relevance of tumoral folylpolyglutamate synthetase and reduced folates for optimal 5-fluorouracil efficacy: experimental data. Eur J Cancer 33: 950–959.
- Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, et al. (1994) Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification. Nat Genet 7: 195–200.
- Zhao M, Li X, Xing C, Zhou B (2013) Association of methylenetetrahydrofolate reductase C677T and A1298C polymorphisms with colorectal cancer risk: a meta-analysis. Biomed Rep 1: 781-791.
- Pereira AC, Schettert IT, Morandini Filho AA, Guerra-Shinohara EM, Krieger JE (2004) Methylenetetrahydrofolate reductase (MTHFR) c677t gene variant modulates the homocysteine folate correlation in a mild folatedeficient population. Clin Chim Acta 340: 99–105.
- Ogino S, Wilson RB (2003) Genotype and haplotype distributions of MTHFR677C>T and 1298A >C single nucleotide polymorphisms: a metaanalysis. J Hum Genet 48: 1–7.
- Crott JW, Mason JB (2005) MTHFR Polymorphisms and colorectal neoplasia. In: Ueland PM, Rozen R, editors. In MTHFR polymorphisms and disease. Texas: Landes Bioscience, pp: 178–196.
- Weisberg I, Tran P, Christensen B, Sibani S, Rozen R (1998) A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab 64: 169–172.
- Girelli D, Friso S, Trabetti E, Olivieri O, Russo C, et al. (1998) Methylenetetrahydrofolate reductase C677T mutation, plasmahomocysteine, and folate in subjects from northern Italy with or without angiographically documented severe coronary atherosclerotic disease: evidence for an important genetic-environmental interaction. Blood 91: 4158–4163.
- Etienne MC, Ilc K, Formento JL, Laurent-Puig P, Formento P, et al. (2004) Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphisms: relationships with 5-fluorouracil sensitivity. Br J Cancer 90: 526–534.
- Etienne MC, Formento JL, Chazal M, Francoual M, Magné N, et al. (2004) Methylenetetrahydrofolate reductase gene polymorphisms and response to fluorouracil-based treatment in advanced colorectal cancer patients. Pharmacogenetics 14: 785–792.
- Xue W, Warshawsky D (2005) Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol Appl Pharmacol 206: 73–93. [Crossref]
- Felton JS, Jagerstad M, Knize MG, Skog K, Wakabayashi K (2000) Contents in foods, beverages, and tobacco. In: Nagao M, Sugimura T (eds.) Food Borne Carcinogens Heterocyclic Amines. John Wiley & Sons Ltd, Chichester, UK, pp: 31–71.
- Turesky RJ (2004) The role of genetic polymorphisms in metabolism of carcinogenic heterocyclic aromatic amines. Curr Drug Metab 5: 169–180.
- Ali-Osman F, Akande O, Antoun G, Mao JX, Buolamwini J (1997) Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. Evidence for differential catalytic activity of the encoded proteins. J Biol Chem 272: 10004–10012.
- Stoehlmacher J, Park DJ, Zhang W, Groshen S, Tsao-Wei DD, et al. (2002) Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst 94: 936-42.
- Chen YC, Tzeng CH, Chen PM, Lin JK, Lin TC, et al. (2010) Influence of GSTP1 I105V polymorphism on cumulative neuropathy and outcome of FOLFOX-4 treatment in Asian patients with colorectal carcinoma. Cancer Sci 101: 530-535.
- Ho V, Massey TE, King WD (2011). Thymidylate synthase gene polymorphisms and markers of DNA methylation capacity. Mol Genet Metab 102: 481-487.
- Marsh S, McKay JA, Cassidy J, McLeod HL (2001) Polymorphism in the thymidylate synthase promoter enhancer region in colorectal cancer. Int J Oncol 19: 383-6. [Crossref]
- Chen J, Hunter DJ, Stampfer MJ, Kyte C, Chan W, et al. (2003) Polymorphism in the thymidylate synthase promoter enhancer region modifies the risk and survival of colorectal cancer. Cancer Epidemiol Biomarkers Prev 12: 958-62.
- Karpinski P, Myszka A, Ramsey D, Misiak B, Gil J, et al (2010) Polymorphisms in methyl-group metabolism genes and risk of sporadic colorectal cancer with relation to the CpG island methylator phenotype. Cancer Epidemiol 34: 338-44. [Crossref]
- Sohn KJ, Croxford R, Yates Z, Lucock M, Kim YI (2004) Effect of the methylenetetrahydrofolate reductase C677T polymorphism on chemo sensitivity of colon and breast cancer cells to 5-fluorouracil and methotrexate. J Natl Cancer Inst 96: 134–44.
- Popat S, Matakidou A, Houlston RS (2004) Thymidylate synthase expression and prognosis in colorectal cancer: a systematic review and meta-analysis. J Clin Oncol 22: 529–536.
- Eddy DM (1990) Screening for colorectal cancer. Ann Intern Med 113: 373-384. [Crossref]
- Johnston PG, Fisher ER, Rockette HE, Fisher B, Wolmark N, et al. (1994) The role of thymidylate synthase expression in prognosis and outcome of adjuvant chemotherapy in patients with rectal cancer. J Clin Oncol 12: 2640–2647. [Crossref]
- Lee KH, Chang HJ, Han SW, Oh DY, Im SA (2013) Pharmacogenetic analysis of adjuvant FOLFOX for Korean patients with colon cancer. Cancer Chemother Pharmacol 71: 843-851.
- Yeh CC, Lai CY, Chang SN, Hsieh LL, Tang R, et al. (2017) Polymorphisms of MTHFR C677T and A1298C associated with survival in patients with colorectal cancer treated with 5-fluorouracil-based chemotherapy Int J Clin Oncol 22: 484-493.
- Chua W, Goldstein D, Lee CK, Dhillon H, Michael M, et al. (2009) Molecular markers of response and toxicity to FOLFOX chemotherapy in metastatic colorectal cancer. Br J Cancer 101: 998-1004. [Crossref]
- Etienne-Grimaldi MC, Milano G, Maindrault-Goebel F, Chibaudel B, Formento JL, et al. (2010) Methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms and FOLFOX response in colorectal cancer patients. Br J Clin Pharmacol 69: 58-66.
- Zhang W, Press OA, Haiman CA, Yang DY, Gordon MA, et al. (2007) Association of methylenetetrahydrofolate reductase gene polymorphisms and sex-specific survival in patients with metastatic colon cancer. J Clin Oncol 25: 3726–3731.
- Ruzzo A, Graziano F, Loupakis F, Rulli E, Canestrari E, Santini D, et al. (2007) Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol 25: 1247–1254.
- Lurje G, Zhang W, Yang D, Groshen S, Hendifar AE, et al. (2008) Thymidylate synthase haplotype is associated with tumor recurrence in stage II and stage III colon cancer. Pharmacogenet Genomics 18: 161–168.
- Helzlsouer KJ, Erlinger TP, Platz EA (2006) C-reactive protein levels and subsequent cancer outcomes: Results from a prospective cohort study. Eur J Cancer 42: 704 – 707.
- GuillemP, Triboulet J (2005) Elevated serumlevels of C-reactive protein are indicative of a poor prognosis in patientswith esophageal cancer. Dis Esophagus 18: 146–50.
- Łukaszewicz-Zając M, Mroczko B, Kozłowski M, Nikliński J, Laudański J, et al. (2012) Higher importance of interleukin 6 than classic tumor markers (carcinoembryonic antigen and squamous cell cancer antigen) in the diagnosis of esophageal cancer patients. Dis Esophagus [Online].
- Fujiwara H, Suchi K, Okamura S, Okamura H, Umehara S, et al. (2011) Elevated serum CRP levels after induction chemoradiotherapy reflect poor treatment response in association with IL-6 in serum and local tumor site in patients with advanced esophageal cancer. J Surg Oncol 103: 62–68. [Crossref]
- Afzal S, Jensen SA, Vainer B, Vogel U, Matsen JP, et al. (2009) MTHFR polymorphisms and 5-FU-based adjuvant chemotherapy in colorectal cancer. Ann Oncol 20: 1660-1666. [Crossref]
- Capitain O, Boisdron-Celle M, Poirier AL, Abadie-Lacourtoisie S, Morel A, et al. (2007) The influence of fluorouracil outcome parameters on tolerance and efficacy in patients with advanced colorectal cancer. Pharmacogenom J 8: 256–267.
- Lecomte T, Ferraz JM, Zinzindohoué F, Loriot MA, Tregouet DA (2004) Thymidylate synthase gene polymorphism predicts toxicity in colorectal cancer patients receiving 5-fluorouracil-based chemotherapy. Clin Cancer Res 10: 5880-5888.
- Schwab M, Zanger UM, Marx C, Schaeffeler E, Klein K, et al. (2008) Role of genetic and nongenetic factors for fluorouracil treatmentrelated severe toxicity: a prospective clinical trial by the German 5-FU toxicity study group. J Clin Oncol 26: 2131–2138.
- Sharma R, Hoskins JM, Rivory LP, Zucknick M, London R, et al. (2008) Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphisms and toxicity to capecitabine in advanced colorectal cancer patients. Clin Cancer Res 14: 817–825.
- Morganti M, Ciantelli M, Giglioni B, Putignano AL, Nobili S, et al. (2005) Relationships between promoter polymorphisms in the thymidylate synthase gene and mRNA levels in colorectal cancers. Eur J Cancer 41: 2176–2183.
- Gusella M, Frigo AC, Bolzonella C, Marinelli R, Barile C, et al. (2009) Predictors of survival and toxicity in patients on adjuvant therapy with 5-fluorouracil for colorectal cancer. Br J Cancer 100: 1549-1557. [Crossref]
- Lai CY, Hsieh LL, Sung FC, Tang R, Bai CH (2013) Tumor site- and stage-specific associations between allelic variants of glutathione S-transferase and DNA-repair genes and overall survival in colorectal cancer patients receiving 5-fluorouracil-based chemotherapy. PLoS One 8: e69039.
- Soyaa SS, Vinoda T, Reddyb KS, Gopalakrishnanc S, Adithana C (2007) Genetic polymorphisms of glutathione-S-transferase genes (GSTM1, GSTT1 and GSTP1) and upper aerodigestive tract cancer risk among smokers, tobacco chewers and alcoholics in an Indian population. Elsevier Europ J Cancer 43; 2698-2706.
- Wei P, Lin S, Chang Y (2011) Cigarette Smoking and Colorectal Cancer: From Epidemiology to Bench Review Article. Journal of Experimental & Clinical Medicine 3: 257-261.


