One of the main problems that immunologists have not solved yet involves the immunosuppression of ongoing exacerbated responses, as in transplant rejection and autoimmune diseases. To overcome these difficulties, different approaches have been used including the administration of immunosuppressive drugs, monoclonal antibodies, and more recently, the utilization of cellular therapy. In this last strategy, huTregs have been proposed as a safe tool to control transplant rejection or collateral immune reactions such as GvHD. Tregs are a subtype of CD4+ T cells characterized by the expression of markers such as CD25, CD127, Foxp3, among others, and their capacity to induce immune tolerance. For this reason, huTregs have become an attractive target for their manipulation and application in the clinic. In this work, we used an established protocol for the expansion of peripheral blood-derived huTregs, adding IL-33 as a putative enhancer of huTregs function, due to its previously identified capacity for driving transplant tolerance. Here, we characterized the phenotype, expansion rate and suppressive function of huTregs33. Although IL-33 did not modify the expression of Tregs canonical markers nor their proliferative activity, it did potentiate their suppressive function. This remarkable observation may be driven in part by the up-regulation of the immune regulatory-related genes, Foxp3, Amphiregulin and ST2. Thus, huTregs33 should be considered for pre-clinical and clinical studies.
Human regulatory T cells (huTregs) have become a target for cellular therapy. In contrast to murine Tregs, huTregs correspond to 5% of Peripheral blood mononuclear cells (PBMC) and express the surface markers CD4, CD25 and low/negative expression of CD127, and transcription factor Forkhead-box P3 (FoxP3), characteristic of murine and human Tregs [1].
Based on their pivotal role in the maintenance of immune tolerance, huTregs isolation and expansion have been improved recently with the purpose of being applied as cellular source of therapy. The most reported protocols include anti-CD3/anti-CD28-coated beads for T cell stimulation, along with administration of human IL-2 and the immunosuppressor drug Rapamycin [2,3], which favors Tregs growth instead of the proliferation of effector CD4+ T. To date, several clinical studies have been performed, and the data indicates that the administration of huTregs is safe even in high doses [4] and seems to ameliorate the symptoms of Graft versus Host Disease (GvHD) in some cases; however, this has not been accomplished in all patients, thus demonstrating the need for more knowledge and treatment alternatives [5-8]. One of the limitations of huTregs effectiveness is the potential lost of their functional stability (or plasticity); in other words, huTregs may convert from a suppressive to an inflammatory phenotype [9]. Among the molecules that could be considered to optimize the production of huTregs, we sought to study IL-33 based on its recent role in the generation of tolerance [10,11]. This cytokine belongs to the IL-1 superfamily of cytokines, and it was initially associated with inflammatory processes such as in asthma, and other CD4+ T helper 2 related diseases, but newer studies have described a role in tissue repair and suppression of organ rejection through the modification of Tregs biology [12,13]. With these antecedents, we decided to use IL-33 as a novel factor for obtaining better and more functional huTregs.
IL-33 does not modify either the phenotype nor cell expansion of human Tregs
With the aim to improve one of the established protocol for purification and expansion of huTregs, we tested the effects of IL-33 on the in-vitro amplification protocol. First, we isolated huTregs from PBMC and cultured them with human IL-2, Rapamycin and IL-33. Freshly isolated huTregs display a canonical phenotype given by high expression of CD25 (~80% of cells), low levels of CD127 (~90% of cells), in addition to high expression of Foxp3 (> 80% of cells), Figure 1A and 1B. In this analysis, we also included the surface marker Nrp1 (because it has been related to Tregs identity) and ST2 or IL-33R (due to the addition of IL-33 in the cultures), obtaining low expression for both of them (< 4% of cells), Figure 1B. In general, all huTregs samples obtained from 5 different donors showed a comparable phenotype with no significant differences in the expression of the molecules analyzed. Next, we cultured huTregs in the presence of human IL-2 and Rapamycin, stimulated with anti-CD3/anti-CD28-coated beads, in the presence (huTregs33) or not of IL-33. huTregs and huTregs33 cells were recovered every 12 days for re-stimulation and media replenishment. At these time points we assessed for cell number and viability, obtaining no differences in total cell number or expansion rate between huTregs and huTregs33 cells, Figure 2A. At the end of the culture (day 36), the phenotype of huTregs and huTregs33 was studied by flow cytometry, obtaining a high percentage of CD4+CD25+CD127low T cells in both conditions (>90%), Figure 2B, which indicates that IL-33 does not change the phenotype of huTregs in the expansion process. Regarding Foxp3, a decrease in the initial level of expression is observed for huTregs and huTregs33 (~70% at day 0 versus ~30% at day 26 for huTregs and ~45% for huTregs33), and for Nrp1 and ST2 expression we did not obtain variations among culture timing or presence of IL-33.
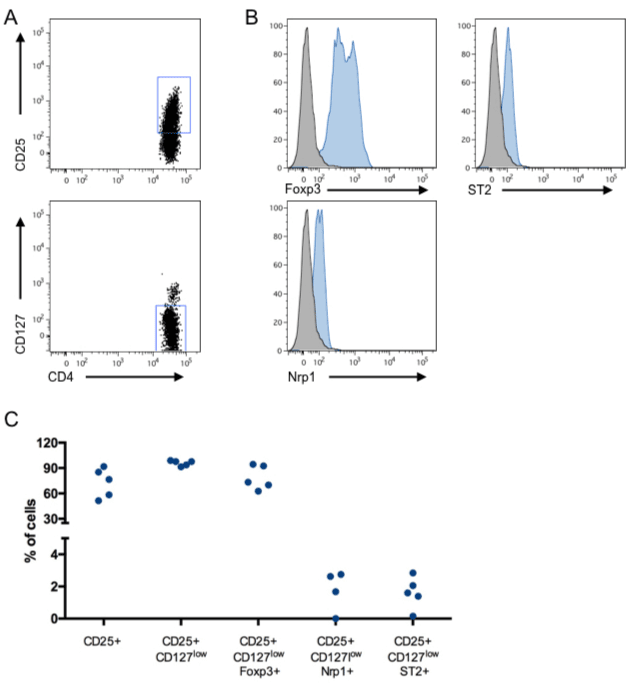
Figure 1. Human regulatory T cells phenotype. Freshly isolated huTregs were stained with antibodies against CD4, CD25, CD127, Foxp3, Nrp1 and ST2 for checking their phenotype using flow cytometry. A. The expression of CD25 on CD4+ T cells is depicted in the top plot, in addition to the expression of CD127 in the gated CD4+CD25+ T cell population, bottom plot. B. The expression of Foxp3, Nrp1 and ST2 was analyzed in the CD4+CD25+CD127low T cell subset. Representative staining of 5 independent donors. C. Pooled data from all samples. n = 5 donors.
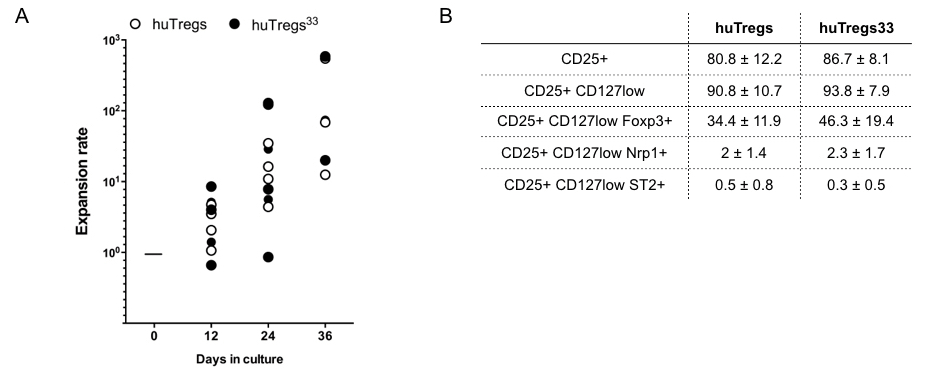
Figure 2. IL-33 does not alter phenotype nor expansion of in vitro cultured huTregs. huTregs isolated from healthy donor´s PBMC were cultured as described in Methods. A. Graph shows the expansion rate of huTregs cultured for 36 days in the presence (filled circle) or not (open circle) of IL-33. B. Table depicting the phenotype of huTregs and huTregs33 evaluated at the end of the culture. The expression of CD4, CD25, CD127, Foxp3, Nrp1 and ST2 was studied by flow cytometry. Values correspond to the mean percentage and the standard deviation. n= 6 different donors.
IL-33 boosts the suppressive function of huTregs
Since the use of IL-33 did not show an apparent effect in huTregs expansion, we sought to evaluate the suppressive function of huTregs. For this purpose, we co-cultured CFSE-labeled CD4+CD25- T cells (or responders) with different ratios of huTregs (or huTregs33) in the presence of polyclonal stimulation by anti-CD3/anti-CD28 beads. After 3 days, cells were recovered and CFSE dilution was studied by flow cytometry. As shown in Figure 3A, ~50% of responders cells proliferated upon anti-CD3/anti-CD28 stimulation, whereas ~25% was observed for responders cells co-cultured with huTregs. Surprisingly, huTregs33 cells inhibited even better than huTregs, bringing responders cells proliferation to only ~15%. This potent inhibitory effect was observed for all ratios tested, Figure 3B. Considering this result, we inferred that IL-33 may influence huTregs function, instead of cell expansion or phenotype, possibly by boosting their cell stability. Thus, we decided to evaluate the expression of certain genes involved in Tregs activity/stability such as Foxp3, ST2 and Amphiregulin (Areg) using quantitative PCR (qPCR), Figure 3C. As mentioned before, Foxp3 has been established as the master transcription factor for Tregs and is involved in the maintenance of the regulatory phenotype and suppressive function [14,15]; ST2 is the receptor for IL-33 and it has been linked with Tregs function in gut mucosa [11,16]; Areg, on the other hand, is an epidermal growth factor that improves Tregs function in vitro and in-vivo mouse models of colitis and tumor, favoring Tregs stability, avoiding conversion toward an inflammatory phenotype [17,18]. For these three genes studied, we observed a clear trend in the up-regulation of their mRNA, although the variations were not significantly different. Based on the results described in this study, we state that the inclusion of IL-33 in the expansion culture for huTregs may improve their suppressive function, possibly by augmenting the expression of relevant genes such as Foxp3, ST2 and Areg.
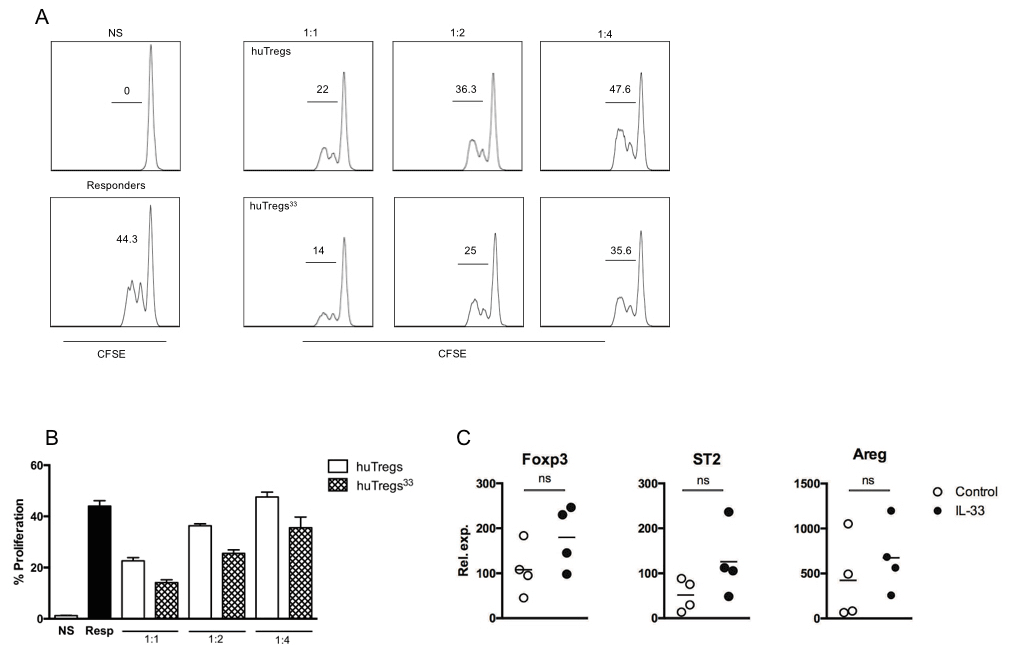
Figure 3. IL-33 produces huTregs with better suppressive activity. huTregs isolated from healthy donor´s PBMC were cultured as described in Methods. At the end of the culture, huTregs or huTregs33 were co-cultured at different ratios with CFSE-labeled CD4+CD25- T cells from the same donor. After 3 days, CFSE dilution was evaluated by flow cytometry. A. Plots depicting CFSE dilution from non-stimulated (NS, top left), activated or responders (bottom left) and CD4+ T cells stimulated in the presence of different ratios of huTregs (top right panel) or huTregs33 (bottom right panel). B. Bar graph showing the percentage of proliferating CD4+ T cells under the indicated conditions (performed in triplicates). Representative data of 3 independent experiments. C. At the end of the expansion protocol, total RNA was isolated from both huTregs (open circles) and huTregs33 (filled circles). Expression of Foxp3, ST2 and Areg was studied by quantitative RT-PCR using HRPT as housekeeping gene. Pooled data from 4 different donors.
Our evidence shows that the adition of IL-33 to huTregs expansion protocol boost the generation of suppressive huTregs33. This observation establishes a new precedent in the improvement of huTregs expansion, which is very significant for use in cellular therapy.
One of the main current problems in transplantation is the inability of assuring the acceptance and survival of the graft. For this purpose several therapies have been developed; however, all these treatments are associated with side effects that harm the patient´s life [19]. Based on what has been described in transplantation clinical trials, the function and application of huTregs are quite promising. Until now, it has been possible to generate protocols for the purification and expansion of huTregs from healthy donors and patients, with the following infusion for treating conditions as transplant rejection, GvHD or autoimmune diseases [1,2,4,20].
Here, we attempted to modify a huTreg expansion protocol with the aim to improve either the rate of huTregs proliferation or the function of the cells. For this intent, we selected IL-33, a cytokine with a wide range of functions, including the ability to induce Tregs13 and enhance their suppressive function [21]. Our findings show that the established markers for huTregs phenotype (CD4+CD25+CD127lowFoxp3+) do not change upon IL-33 treatment, obtaining a phenotype for huTregs33 similar to control cells. However, when we performed a suppression assay, huTregs33 were more suppressive than control huTregs. This remarkable finding gives us the idea that even if IL-33 is not changing the phenotype of huTregs (at least not modifying the expression of CD25, CD127, Nrp1 or ST2), IL-33 may be targeting other mechanisms in the cell, resulting in huTregs33 becoming better suppressors. In the process of Tregs differentiation, the concepts of stability and/or plasticity have been recognized since Tregs facing an inflammatory milieu may become unstable, down-regulating the expression of Foxp3 [22,23], or they may convert to inflammatory cells by gaining a Th17-like phenotype [24]. In this regard, molecules such as ST2 and Areg have been implicated in the maintenance of Tregs phenotype and function. For instance, the receptor for IL-33, or ST2, has served as a marker for highly activated murine Tregs with a Th2-like phenotype [25], and also for suppressive Tregs residing in the intestine [11]. Conversely, the treatment with IL-33 (for 24 hours) of freshly isolated huTregs from diabetic patients has been tested, resulting in an increased ST2 expression [26], no variation in Foxp3 levels, but greater capacity to block IFN-g expression on effector T cells [20]. Comparing this last study with ours, it seems surprising that IL-33 only improves huTregs function from diabetic, and not healthy donors (as in this current report). Also, it is not reported whether IL-33-treated diabetic huTregs inhibit effector T cell proliferation. We believe that age of PBMC donors and in-vitro conditions (media, activation and duration of the cell culture) may affect the biology of the final huTreg population. Following the same line of what is reported for ST2, Areg seems to enhance the suppressive activity of tissue-resident Tregs as shown in mouse models of carcinoma and viral infection [27,28]. Supporting our findings, Carney et al. reported that anti-CD3 activated huTregs up-regulate Areg at the gene and protein levels [18], and even more ensuring are the observations stating that IL-33 is involved in Areg synthesis [29,30].
Taking all of this together, we think that the inclusion of IL-33 in the expansion protocol for human Tregs is a novel improvement for obtaining up-graded regulatory T cells, which may ease the manufacture and administration of these cells in the clinical setting.
Isolation and expansion of Human Regulatory T cells
Blood samples were obtained from healthy donors recruited at the Blood Bank of Hospital Militar de Santiago (La Reina, Chile) and Blood Bank of Clinica Santa Maria (Providencia, Chile). The blood was processed to obtain PBMC by Ficoll-Paque density gradient. Cells were recovered and washed twice with 1X PBS for further huTregs isolation using the CD4+CD25+ T regulatory cells isolation kit from Miltenyi (California, USA). For huTregs culture and expansion we followed a protocol from Safinia et al., 2016. Briefly, 1x106 cells were stimulated with anti-CD3/anti-CD28 beads (Invitrogen, USA) and cultured in X-VIVO 15 media (Lonza, Basel, Switzerland) complemented with 5% of AB+ plasma, 500 U/ml of human IL-2 (Miltenyi Biotech) and 100nM Rapamycin (SigmaAldrich). Addicionally, half of the sample was stimulated with 20 ng/ml of rhIL-33 (Peprotech). Media including fresh IL-2 and Rapamycin, was replaced every 2 days, antiCD3/anti-CD28 beads were replenished every 12 days. At day 36, expanded huTregs were recovered, counted and analyzed for phenotype and function.
Flow cytometry
huTregs phenotype was analyzed by flow cytometry using anti-human CD4 (clone SK3), CD25 (clone BC96), CD127 (clone A0195), Foxp3 (clone 206D, all from BioLegend, San Diego, CA), Nrp1 (clone 446921) and ST2 (clone not provided, R&D Systems, Minneapolis, MN). All antibodies were conjugated with FITC, PE, PerCP, APC, PeCy7 or Pacific Blue. FACS data acquisition was performed with FACSCanto II (Beckton Dickinson, Franklin Lakes, NJ), using the Diva software (BD Biosciences, San Jose, CA), and analyzed with FlowJo software (Tree Star, OH, USA).
Suppression assay in vitro
1x105 CFSE-labeled CD4+CD25- T cells were cultured with anti-CD3/anti-CD28 beads (Dynabead human Treg Expander, Life Technologies, MA, USA) at a 1:20 ratio (bead: T cell). 1x105, 5 x 104 and 2.5x104 huTreg cells were added into culture. At day 3 cells were harvested, the supernatant recovered and the cells stained with CD4-PECy7 for flow cytometry analysis.
qRT-PCR analysis
RNA from in vitro expanded huTreg cells was extracted using the RNeasy® Mini Kit (Qiagen, Hilden, Germany). cDNA samples were prepared using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). The expression of Foxp3, Nrp1, ST2, Areg and hypoxanthine-guanine phosphoribosyl transferasa (HPRT) was quantified using the Mx3000P quantitative PCR system (Agilent Technologies, Santa Clara, CA), using 5X HOT FIREPol EvaGreen qPCR Supermix (Solis BioDyne, Tartu, Estonia) as fluorescence detector. Primer sequences were: Foxp3, F: CCT ACC CAC TGC TGG CAA AT, R: AAG CCT TGG TCA GTG CCA TT; Nrp1, F: AGC ACC GAG AGA ACA AGG TG, R: CAC CTG TGA GCT GGA AGT CAT; ST2, F: TTC TGG ATT GAG GCC ACT CTG, R: CCA GGT AGC ATA TCT CTC CCA; Areg, F: GAC ACC TAC TCT GGG AAG CG, R: GTA GTC ATA GTC GGC TCC CG; and HPRT, F: CAA GCT TGC TGG TGA AAA GGA C, R: GTC AAG GGC ATA TCC TAC AAC AAA.
Statistical analysis
Data were analyzed using an unpaired Student’s t-test or a Mann–Whitney test (two-tailed). In all cases, P < 0.05 was considered with statistical significance. For data analysis, GraphPad Prism v5.0 (GraphPad Software, CA, USA) was used.
We are grateful to Dr. Giovanna Lombardi and Dr. Niloufar Safinia (Kings College London, London, UK) for scientific advice with the protocol for human Tregs expansion; to Dr. Francisca Alcayaga (Cells for Cells, Chile) for facilitating certain reagents; and to Ms. Petra Sergent (Darmouth College, NH, USA) for reading and editing the manuscript.
This work was supported by PMI/UAN 1301 and FONDECYT Regular 1160347 grants.
The authors declare no conflict of interest.
- Trzonkowski P, Bacchetta R, Battaglia M, Berglund D, Bohnenkamp HR, et al. (2015) Hurdles in therapy with regulatory T cells. Sci Transl Med 7: 304ps18. [Crossref]
- Strauss L, Whiteside TL, Knights A, Bergmann C, Knuth A, et al. (2007) Selective survival of naturally occurring human CD4+CD25+Foxp3+ regulatory T cells cultured with rapamycin. J Immunol 178: 320-329. [Crossref]
- Scottà C (2015) Impact of immunosuppressive drugs on the therapeutic efficacy of ex vivo expanded human regulatory T cells. Haematologica 101: 91-100.
- Bluestone (2015) Type 1 diabetes immunotherapy using polyclonal regulatory T cells. Sci Transl Med 7: 315ra189.
- Safinia N (2016) Successful expansion of functional and stable regulatory T cells for immunotherapy in liver transplantation. Oncotarget 7: 7563-77.
- Trzonkowski P, Bieniaszewska M, JuÅ›ciÅska J, Dobyszuk A, Krzystyniak A, et al. (2009) First-in-man clinical results of the treatment of patients with graft versus host disease with human ex vivo expanded CD4+CD25+CD127- T regulatory cells. Clin Immunol 133: 22-26. [Crossref]
- Brunstein CG (2012021 Copyright OAT. All rights reservulatory cells in adults transplanted with umbilical cord blood?: safety profile and detection kinetics Infusion of ex vivo expanded T regulatory cells in adults transplanted with umbilical cord blood?: safety profile and. Blood 117: 1061–1070.
- Di Ianni M (2011) Tregs prevent GVHD and promote immune reconstitution in HLA-haploidentical transplantation. Blood 117: 3921-3928.
- Safinia N, Scotta C, Vaikunthanathan T, Lechler RI, Lombardi G (2015) Regulatory T cells: Serious contenders in the promise for immunological tolerance in transplantation. Front Immunol 6: 1-16.
- Gajardo T, Morales R A, Campos-Mora M, Campos-Acuña J, Pino-Lagos K (2015) Exogenous interleukin-33 targets myeloid-derived suppressor cells and generates periphery-induced Foxp3+ regulatory T cells in skin-transplanted mice. Immunology 146.
- Schiering C, Krausgruber T, Chomka A, Fröhlich A, Adelmann K, et al. (2014) The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513: 564-568. [Crossref]
- Yin H (2013) IL-33 accelerates cutaneous wound healing involved in upregulation of alternatively activated macrophages. Mol Immunol 56: 347-353.
- Carrasco TG (2015) Alarmin’ immunologists: IL-33 as a putative target for modulating T cell-dependent responses. Front Immunol 6: 4-11.
- Wood KJ, Bushell A, Hester J (2012) Regulatory immune cells in transplantation. Nat Rev Immunol 12: 417-430. [Crossref]
- Bennett CL (2001) The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet 27: 20-21 (2001).
- Schmidt A, Eriksson M, Shang MM, Weyd H, Tegnér J (2016) Comparative Analysis of Protocols to Induce Human CD4+Foxp3+ Regulatory T Cells by Combinations of IL-2, TGF-beta, Retinoic Acid, Rapamycin and Butyrate. PLoS One 11: e0148474. [Crossref]
- Zaiss DMW (2014) Amphiregulin enhances regulatory T cell suppressive function via the epidermal growth factor receptor Dietmar. Immunity 38: 275-284.
- Carney K, Chang YR, Wilson S, Calnan C, Reddy PS, et al. (2016) Regulatory T-cell-intrinsic amphiregulin is dispensable for suppressive function. J Allergy Clin Immunol 137: 1907-1909. [Crossref]
- Chinen J, Buckley RH (2011) NIH Public Access Transplantation immunology: Solid Organ and bone marrow. J Allergy Clin Immunol 125: 1-26.
- Ryba-Stanisławowska M, Buksa L, Brandt A, Juhas U, Myśliwiec M (2017) IL-33 improves the suppressive potential of regulatory T cells in patients with type 1 diabetes. Diabetes Res Clin Pract 128: 67-73. [Crossref]
- Turnquist HR (2011) IL-33 expands suppressive CD11b+ Gr-1(int) and regulatory T cells, including ST2L+ Foxp3+ cells, and mediates regulatory T cell-dependent promotion of cardiac allograft survival. J Immunol 187: 4598–610.
- McClymont SA, Putnam AL, Lee MR, Esensten JH, Liu W, et al. (2011) Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunol 186: 3918-3926. [Crossref]
- Sakaguchi S, Vignali DA, Rudensky AY, Niec RE, Waldmann H (2013) The plasticity and stability of regulatory T cells. Nat Rev Immunol 13: 461-467. [Crossref]
- Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh-hora M, et al. (2014) Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat Med 20: 62-68. [Crossref]
- Siede J (2016) IL-33 receptor-expressing regulatory t cells are highly activated, Th2 biased and suppress CD4 T Cell proliferation through IL-10 and TGF?? Release. PLoS One 11: 1-15.
- Ryba-Stanisławowska M, Werner P, Skrzypkowska M, Brandt A, Myśliwska J (2016) IL-33 Effect on Quantitative Changes of CD4+CD25highFOXP3+ Regulatory T Cells in Children with Type 1 Diabetes. Mediators Inflamm 2016: 9429760. [Crossref]
- Yuan CH (2015) Amphiregulin activates regulatory T lymphocytes and suppresses CD8+ T cell-mediated anti-tumor response in hepatocellular carcinoma cells. Oncotarget 6.
- Dai K, Huang L, Chen J, Yang L, Gong Z (2014) Amphiregulin promotes the immunosuppressive activity of intrahepatic CD4 + regulatory T cells to impair CD8 + T cell immunity against hepatitis B virus infection. Immunology 144, n/a-n/a.
- Monticelli LA (2015) IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc. Natl. Acad. Sci. U. S. A. 112: 10762-10767.
- Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, et al. (2013) A special population of regulatory T cells potentiates muscle repair. Cell 155: 1282-1295. [Crossref]



