Abstract
It has been previously reported that G1 arrest after gamma irradiation is suppressed by inhibitor of poly (ADP-ribose) polymerase (Parp1), 3-aminobenzamide (3-AB) in C3D2F1 3T3-a cells, which has no mutation in exon 5 to exon 9 of the p53 gene. To elucidate the mechanisms that Parp1 is involved in G1 arrest through the p53 pathways after gamma irradiation, we investigated the effect of 3-AB on downstream following p53 protein accumulation in the p53-dependent signaling pathway at G1 arrest. The transactivation activity of p53 was assessed by the binding activity of p53 to its consensus binding sequence by gel shift assay. The expression of Waf1/Cip1/p21 and Mdm2 mRNA was analyzed by Northern blot. The DNA binding activity of p53 after gamma irradiation was increased dose dependent manner and moreover the increase of the activity was enhanced in the presence of 3-AB. The expression of Waf1/Cip1/p21 and Mdm2, which are downstream factors of p53, was induced by approximately 8- and 12-fold at 2.5 h after 8 Gy irradiation in the absence of 3-AB, respectively. These expressions were suppressed in the presence of 3-AB. In present study, the possibility has been shown that Parp1 participates in the regulation of Waf1/Cip1/p21 and Mdm2, which are transcriptionally activated by p53, suggesting that Parp1 is involved in the downstream of p53 dependent signal transduction after DNA damage.
Key words
Parp1, DNA damage, G1 arrest, p53, p21, Mdm2
Introduction
Multiple lines of evidence point to poly (ADP-ribose) polymerase (Parp1) being involved in cellular DNA damage repair. Cytotoxicity induced by DNA damaging factors like gamma irradiation is worsened in the presence of the Parp inhibitor 3-aminobenzamide (3-AB). DNA repair in response to alkylating agents is inhibited in cells expressing low levels of Parp1 or in dominant negative Parp1 mutants that only express the DNA binding domain; and inhibition of Parp1 expression with anti-sense RNA against Parp1 mRNA delays DNA repair [1-3]. The role of Parp1 in DNA repair has been reported in cell-free systems as well; the DNA repair process is temporarily interrupted in a Parp1 dependent manner in the presence of NAD [4]. Since Parp1 specifically recognizes the cleaved DNA terminus, it is thought to function in the DNA excision repair pathway associated with strand breakage. In addition to the evidence suggesting that Parp1 works directly on DNA repair, there are reports stating that Parp1 greatly promotes the activity of DNA ligase in chromatin [5]. In addition, the “histone-shuttling model" states that poly-ADP-ribosylated histones lose their affinity for DNA and promote DNA repair by loosening the structure around the DNA cleavage end [6]. Thus, Parp1 appears to be involved in various physiological functions associated with DNA strand breaks.
Protein levels of the transcription factor p53 are increased in cells treated with DNA damaging agents by a post-translational mechanism that prolongs the half-life of p53 in cells [7,8]. This prolonged half-life and accumulation of p53 in the nuclei affects expression of genes harboring p53 binding sites in the promoter regions, leading to changes in gene expression and induction of G1 arrest [7-9]. Of the many genes known to be regulated by p53, the induction of Waf1/Cip1/p21, following p53 stabilization suppresses the activities of G1 cyclin and the catalytic enzyme of the cyclin dependent kinase (Cdk) complex [10,11]. In addition, p53 protein also induces the expression of other genes such as Mdm2 and Gadd45 that may be important for functions other than the induction of cell cycle arrest following DNA damage [9,12,13]. It has been suggested that the causative gene product of the disease Ataxia Telangiectasia (AT) is involved in the signaling pathway from DNA damage in increasing p53 protein levels, whereas B-lymphocytes AT patients exhibited no increase in the accumulation of p53 protein after DNA damage [14].
In the present study, the possibility of Parp1 as the key molecule involved in transmitting the quantitative information of DNA damage to p53 has been investigated. To elucidate the hypothesis, we analyzed the involvement of Parp1 in the downstream of p53 signaling pathway after DNA damage by measuring DNA binding and transcription levels of p53 regulated genes in the presence of the Parp1 inhibitor 3-AB. These analyses reveal a role for Parp1 in the G1 arrest following gamma irradiation but require further elucidation of the pathways through which this signal is transmitted.
Materials and methods
Cell culture
C3D2F1 3T3-a cell line, a fibroblast cell line that was established from 14-day-old embryos of C3H/HeJ and DBA/2J mice, was used in this study [15]. The cells were established and provided by Professor Katsuhiro Ogawa and his collaborators at Asahikawa Medical University. Cells were seeded at 3 × 105 cells per 10-cm dish and cultured in DMEM (ICN Biochemical Inc. Costa Mesa, CA, USA) containing 10% fetal bovine serum (FBS). The cells were passaged every 3 days. The doubling time was approximately 17 hours. This cell line has been reported to have no mutation in exon 5 to exon 9 of the p53 gene [15]. Gamma-irradiation of C3D2F1 3T3-a cells were carried out using 60Co gamma-irradiator at 1 Gy/min.
Inhibitor of the Parp
3-AB was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan).
Gel shift assay
Upon stabilization, p53 regulates the transcription of genes having p53 recognition sequences. As an index for measuring the transcriptional activation ability of intracellular p53, a gel shift assay that measures its binding activity towards an oligo-DNA probe having p53 recognition sequences is used. As a p53 binding consensus sequence, the self-complementary double stranded DNA 5'-GGACATGCCCGGGCATGTCC-3' has been reported [16]. The 20-mer DNA was synthesized, heated in an annealing buffer solution [20 mM Tris-Cl (pH 7.5), 10 mM MgCl2, 0.1 M NaCl] at 95°C for 10 min and further at 65°C for 90 min, cooled until room temperature, and annealed. This DNA was isolated by 15% polyacrylamide gel electrophoresis. The obtained double-stranded DNA fragment was eluted with 2 mL of 0.2 M triethylammonium bicarbonate by shaking overnight at 4°C, collected by ethanol precipitation, and dissolved in 30 mM NaCl, 10 mM Tris-Cl (pH 8.0), 1 mM EDTA. This was labeled by 5'-end kination using T4 polynucleotide kinase and [γ-32P] ATP. Thereafter, a nuclear extract was prepared according to the method of Funk et al. First, the cells were washed twice with phosphate-buffered saline (PBS), and 2 mL of buffer solution A [20 mM potassium-HEPES (pH 7.6), 20% glycerol, 10 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1 % Triton X-100, 1 mM PMSF, 1 mM DTT] was poured per 10-cm dish. After detaching the cells from the dish, they were transferred to a centrifuge tube and allowed to stand on ice for 5 min. The cells were agitated 20 times with a Dounce-type homogenizer (L pestle) and centrifuged again at 250 × g for 5 min to obtain the nuclear fraction as a precipitate. To this, 2 mL of buffer solution A containing 0.5 M NaCl was added and the precipitate extracted by shaking at 4°C for 30 min. Further, the supernatant was collected by centrifugation at 10,800 × g for 15 min. The supernatant was dialyzed against buffer solution A and centrifuged at 250 × g for 5 min to remove precipitate. Protein quantification was carried out using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA, USA), and nuclear extracts were dispensed and stored at -80°C. The reaction was carried out for 10 min at 25°C in buffer solution A containing 150 mM NaCl, by adding 3 × 104 cpm/0.6 mg of 32P-labeled probe, 4 mg of poly(dI-dC), and 50 ng of anti-p53 antibody (Ab421, Merck Millipore, Darmstadt, Germany) to the nuclear extract (equivalent to 2 mg of protein), bring the volume to 18.5 µL. Subsequently, 4% polyacrylamide gel electrophoresis was carried out for 4 hrs at 120 V and 4°C after adding 1.9 µL of a solution containing 5 % glycerol, 50 mM EDTA, 0.05 % bromophenol blue, and 0.05 % xylene cyanol. The gel was dried and analyzed with an image analyzer (BAS2000, FUJIFIRM, Tokyo, Japan).
Northern blot analysis
From the N-terminal translation initiation codon of mouse Waf1/Cip1/p21 cDNA, the following two oligonucleotides namely, a 64-mer and a 62-mer oligonucleotides, sense 5'-ATGTCCAATCCTGGTGATGTCCGACCTGTTCCGCACAGGAGCAAAGTGTGCCGTTGTCTC-3'(64-mer) and antisense 5'-ACGGCAACAGAGAAGCCAGGGCACCTGTCACTCGTCAACTCGGCACTAACGCTACGCGAGTA-3'(62-mer) were set in 110 bases [17] and prepared by the solid phase phosphoramidite method using the Applied Biosystems 392 DNA/RNA synthesizer. After heating 150 ng each of these two oligonucleotides at 95°C for 5 min and 65°C for 2 min, they were gradually cooled to room temperature and annealing was carried out. Thereafter, it was labeled with DNA polymerase I (Klenow fragment) by an extension reaction using [α-32P]dCTP. A region having 100% homology between human and mouse was used as a primer for Mdm2 [18]. In other words, a sense primer 5'-TGTGCAATACCAACATGTCTG-3' (21-mer) and an anti-sense primer 5'-TTCCAATAGTCAGCTAAGG-3' (19-mer) were defined for amino acids 1 to 7 and amino acids 298 to 304 respectively of the human MDM2 cDNA and isolated using the Reverse-Transcriptase Polymerase Chain Reaction (RT-PCR) method. The 0.8-kbp HindIII and SacI fragments sub-cloned into pBluescript SK(+) vector (Stratagene, La Jolla, CA, USA) were used as probes. With 50 ng of DNA labeled using the multi-prime method as the template, [α-32P]dCTP labeling was carried out using the Megaprime DNA labeling system (Amersham, BUCKS, UK ) and using the Sephadex G-50 (Pharmacia Biotech, Uppsala, Sweden) spin column method, the free nucleotides were excluded leaving labeled probes. The specific activity was 1.0 x 105 cpm/ng.
Results
Effect of 3-AB on p53 transcriptional activity after gamma irradiation
The effect of 3-AB on the downstream of p53 accumulation stage in the G1 arrest signaling pathway after DNA damage was analyzed using the electrophoretic gel shift assay. In the nuclear extracts derived from C3D2F1 3T3-a cells, the DNA binding activity of p53 was detected. In accordance with the method of Zauberman et al. [19] the antibody Ab421 against the C-terminus of p53 protein, which is known not to interfere the specific DNA binding activity of p53, was added in the reaction mixture, and the p53 protein-DNA complex band was further super-shifted to obtain a clearer signal [19]. To compare to the signal, purified human p53 protein expressed using a baculovirus expression system was used as a control (Figure 1A). The binding activity of p53 was observed with the dose dependence of gamma rays. The binding activity 1 hour after 12 Gy irradiation was 6-times higher than that without irradiation (Figure 1B). In the presence of 3-AB at 4 mM, we noted that basal DNA binding activity of p53 protein is slightly increased. In the presence of 3-AB, the increase in the DNA binding activity of p53 was further enhanced 1 hour after 8 Gy irradiation (Figure 2), suggesting the 3-AB did not inhibit the p53 DNA binding and rather it enhanced.
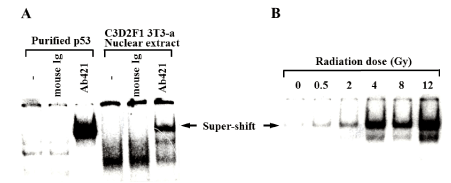
Figure 1. Specific DNA binding activity of p53 protein after gamma-irradiation
The specific DNA binding activity of p53 to DNA one hour after gamma irradiation is shown by gel shift assay. (A) Purified human p53 protein and the nuclear extract solution of C3D2F1 3T3-a cells were prepared. Arrows indicate the bands super-shifted by the p53 antibody (Ab421). (B) The specific DNA binding activity of nuclear extract p53 following irradiation 1 hour after gamma irradiation doses of 0, 0.5, 2, 4, 8, and 12 Gy is shown

Figure 2. Effect of 3-AB on the specific DNA binding activity of p53 protein after gamma-irradiation
The effect of 4 mM 3-AB addition one hour after 8 Gy irradiation in the specific binding activity of p53 to DNA is shown. The arrow indicates the bands super-shifted by the p53 antibody (Ab421)
Effect of 3-AB on mRNA expression levels of WAF1/CIP1/p21 and Mdm2
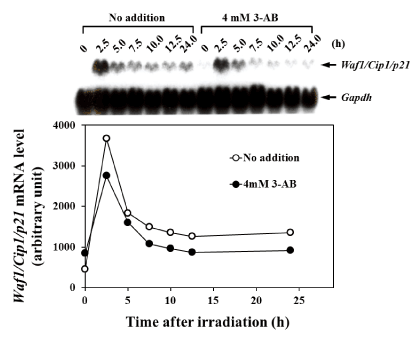
Figure 3. Effect of 3-AB on the induction of Waf1/Cip1/p21 mRNA expression after gamma-irradiation
The effect of the addition of 4 mM 3-AB on the induction of Waf1/Cip1/p21 m RNA expression at 0, 2.5, 5.0, 7.5, 10.0, 12.5, and 24 hours after gamma-irradiation with 8 Gy is shown. The upper panel shows the northern blot analysis of mRNA levels at each time point. In the lower section, the intensity of bands for each time-duration is plotted. The white circles indicate samples that have not been treated with 3-AB, and the black circles indicate samples that have been treated with 4 mM 3-AB. The vertical axis shows the relative expression levels of Waf1/Cip1/p21 mRNA, and the horizontal axis shows the time after gamma-irradiation. The data were normalized using Gapdh as an endogenous control
Effect of 3-AB on mRNA expression levels of p53-responsive signaling factors was analyzed. In C3D2F1 3T3-a cells, the mRNA expression level of WAF1/CIP1/p21 transiently increased about 8 times 2.5 h after 8 Gy irradiation and gradually decreased over 5 h of irradiation. The expression was sustained to some extent even after 12.5 h and 24 h, when G1 arrest became noticeable. However, in the presence of 4 mM 3-AB, the expression of WAF1/CIP/p21 mRNA after 2.5 h was suppressed by approximately 30%, and thereafter it was suppressed by about 50% compared to non-addition condition of 3-AB (Figure 3). In C3D2F1 3T3-a cells, the expression of Mdm2 mRNA transiently increased to about 12 times 2.5 h after 8 Gy irradiation. Subsequently, it’s level decreased to about 50% in 5 h, compared to the peak at 2.5 h after irradiation and the Mdm2 expression gradually decreased thereafter. In the presence of 4 mM 3-AB, the increase of Mdm2 expression was suppressed by about 75% 2.5 h after irradiation. Of note, continuous higher Mdm2 expression was observed until 24 h after irradiation when the cells were in G1 arrest (Figure 4). This higher Mdm2 expression at 10-24 hr in the presence of 3-AB may be related to higher p53 DNA binding activity in the presence of 3-AB shown in Figure 2.
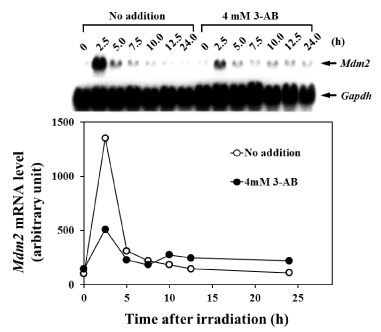
Figure 4. The effect of 3-AB on the induction of Mdm2 mRNA expression after gamma irradiation
The effect of the addition of 4 mM 3-AB on the induction of mdm2 mRNA expression at 0, 2.5, 5.0, 7.5, 10.0, 12.5, and 24 hours after gamma irradiation with 8 Gy is shown. The upper part shows the northern blot analysis of Mdm2 mRNA levels. In the lower part, the intensity of the bands for each time-point is plotted. The white circles indicate the controls without the addition of 3-AB and the black circles indicate the addition of 4 mM 3-AB. The vertical axis shows the relative expression levels of mdm2 mRNA and the horizontal axis shows the time after gamma-irradiation. The data were normalized using Gapdh as an endogenous control
Discussion
There are many reports involving the G1 arrest mechanism after DNA damage. After DNA damage, the stabilization and intracellular accumulation of p53 protein occurs. Then its transcriptional activation ability increases, and it acts as a transcriptional regulator to induce the expression of proteins that suppress the activities of G1 cyclin-Cdk complex, leading to G1 arrest [11]. In C3D2F1 3T3-a cells, after gamma-irradiation, G1 arrest is suppressed by the Parp inhibitor, 3-AB, and G2-phase arrest is accelerated [20]. Given this, we examined the effect of 3-AB on the p53-dependent G1 arrest signaling pathway. It is known that after DNA damage, the binding activity of p53 protein towards the specific DNA recognition sequence increases. In the presence of 3-AB at 4 mM, we observed that basal DNA binding activity of p53 protein is slightly increased. The addition of 3-AB further increased in the DNA binding activity of p53 protein after gamma-irradiation at 8 Gy. Therefore, the poly (ADP-ribose) synthesis reaction may be involved in the p53 DNA binding step. However, the p53 protein recognition consensus DNA sequence used was only 20 base pairs, whereas in a physiological context it is likely that additional regulatory regions exist in the promoter sequences of p53 target genes and that p53 interacts with other transcription factors, both of which can affect p53 DNA binding. Therefore, this DNA binding assay alone cannot conclusively determine the role of Parp1 in the transcriptional activation ability of p53. We examined the influence of 3-AB on the mRNA expression levels of Waf1/Cip1/p21 and Mdm2, two genes that are known to be induced by p53 following gamma-irradiation.
WAF1/CIP1/p21 has been isolated and analyzed as an inhibitor of cyclin dependent kinase. The presence of p53 recognition sequences in the region upstream of the human Waf1/Cip1/p21 gene has been reported [17], and induction of mRNA expression is observed at about 2 h after gamma-ray irradiation [11]. MDM2 forms a complex with p53 protein and it is known as a protein that negatively regulates the function of p53 as a ubiquitin ligase [21]. Two p53 binding sequences are present upstream of the Mdm2 gene [22], and the expression of mRNA is very strongly induced by p53 protein after p53 stabilization following gamma-irradiation [9]. In this present study, the increase in the transient expression of mRNA was observed in the Waf1/Cip1/p21 and Mdm2 genes 2.5 h after gamma irradiation with 8 Gy. The transient increase in expression of the both mRNA was reduced in the presence of 3-AB. Thus, the inhibition of Parp1 activity was shown to antagonize in the radiation dependent increase in mRNA levels of these genes. The inhibitory effect at this early time point varied for the two transcripts and was more clearly observed for Mdm2 mRNA. This may be due to differences in the contribution of p53 protein as a transcription factor in the regulatory region of each gene. Under the same experimental conditions, even at 12 h after irradiation when G1 arrest was observed, expression of the mRNA of Waf1/Cip1/p21 genes was continuously suppressed to about 50% in the presence of 3-AB. The continuous suppression of the expression level of genes directly involved in cell cycle progression, through the inhibition of cyclin-dependent kinase, is considered an important factor that causes the suppression of G1 arrest in the presence of 3-AB. On the contrary, Mdm2 mRNA expression after 10 h of gamma-irradiation was about two-times higher in the presence of 3-AB than that in its absence. Kastan et al. reported that over-expression of Mdm2 suppressed G1 arrest after gamma-irradiation [18]. Therefore, the increase in Mdm2 expression levels after 10 h of gamma-irradiation may also contribute to the G1 arrest suppression seen following 3-AB treatment. Since Mdm2 is a repressor of p53 protein, Mdm2 functions to terminate the G1 arrest [23], and there has been no evidence for its involvement in the induction of G1 arrest. However, this study has indicated that the transient expression of Mdm2 may be involved for the induction of G1 arrest. In order to confirm the importance of changes in expression of these mRNAs as a factor for G1 arrest suppression in the presence of 3-AB, it is necessary to examine whether the changes in expression of Waf1/Cip1/p21 and Mdm2 also occur at the protein level.
The results gleaned from our analysis of p53 binding and transcriptional regulation following 3-AB treatment suggest that Parp1 is involved in the transcriptional activation and regulation of Waf1/Cip1/p21 and Mdm2 involving the p53-dependent signaling pathway after DNA damage. In mouse C3D2F1 3T3-a fibroblast cells. In human MCF-7 cells and BJ/TERT cells, another Parp inhibitor 1,5-dihydroxyisoquinoline also suppressed Waf1/Cip1/p21 and Mdm2 mRNA expression after gamma irradiation, suggesting that a conserved role of Parp1 in G1 arrest regulation after gamma-irradiation [24].
The changes in mRNA expression by 3-AB could be due to the alteration of mRNA half-life or changes in the p53 transcriptional activation ability for mRNA expression. These possibilities can be examined by measuring the half-life of each mRNA and studying the transcriptional activation ability of each gene by nuclear run-on assays. There is also a possibility that the poly (ADP-ribose) synthesis reaction induces G1 arrest by directly inhibiting the activity of cyclin-dependent kinase or the activity of the target protein of the downstream cyclin-dependent kinase. Alternatively, a p53-independent G1 arrest mechanism could exist, which involves cross-talk with the p53-dependent pathway. Therefore, it is necessary to consider the possibility of Parp1 involvement in this alternative pathway. It is considered that Parp1 likely functions promptly as a signal of DNA strand breakage and accurately transmits quantitative information regarding DNA strand breakage.
Conflicts of interest
The authors declare that there is no conflicts of interest.
Acknowledgement
This work was supported in part by MEXT KAKENHI (15K14416).
References
- Nduka N, Skidmore CJ, Shall S (1980) The enhancement of cytotoxicity of N-methyl-N-nitrosourea and of gamma-radiation by inhibitors of poly(ADP-ribose) polymerase. Eur J Biochem 105: 525-530. [Crossref]
- Molinete M, Vermeulen W, Burkel A, Menissier-de Murcia J, Kupper JH, et al. (1993) Overproduction of the poly(ADP-ribose) polymerase DNA-binding domain blocks alkylation-induced DNA repair synthesis in mammalian cells. EMBO J 12: 2109-2117.
- Ding R, Pommier Y, Kang VH, Smulson M (1992) Depletion of poly(ADP-ribose) polymerase by antisense RNA expression results in a delay in DNA strand break rejoining. J Biol Chem 267: 12804-12812. [Crossref]
- Satoh MS, Lindahl T (1992) Role of poly(ADP-ribose) formation in DNA repair. Nature 356: 356-358. [Crossref]
- Creissen D, Shall S (1982) Regulation of DNA ligase activity by poly(ADP-ribose). Nature 296: 271-272. [Crossref]
- Realini CA, Althaus FR (1992) Histone shuttling by poly(ADP-ribosylation). J Biol Chem 267: 18858-18865. [Crossref]
- Tishler RB, Calderwood SK, Coleman CN, Price BD (1993) Increases in sequence specific DNA binding by p53 following treatment with chemotherapeutic and DNA damaging agents. Cancer Res 53: 2212-2216. [Crossref]
2021 Copyright OAT. All rights reserv
- Maltzman W, Czyzyk L, (1984) UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol 4: 1689-1694.
- Price BD, Park SJ (1994) DNA damage increases the levels of MDM2 messenger RNA in wtp53 human cells. Cancer Res 54: 896-899. [Crossref]
- Deb SP, Munoz RM, Brown DR, Subler MA, Deb S (1994) Wild-type human p53 activates the human epidermal growth factor receptor promoter. Oncogene 9: 1341-1349.
- el-Deiry WS, Harper JW, O'Connor PM, Velculescu VE, Canman CE, et al. (1994) WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res 54: 1169-1174. [Crossref]
- Perry ME, Piette J, Zawadzki JA, Harvey D, Levine AJ (1993) The mdm-2 gene is induced in response to UV light in a p53-dependent manner. Proc Natl Acad Sci U S A 90: 11623-11627. [Crossref]
- Zambetti GP, Bargonetti J, Walker K, Prives C, Levine AJ (1992) Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev 6: 1143-1152.
- Kastan MB, Zhan Q, El-Deiry WS, Carrier F, Jacks T, et al. (1992) A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71: 587-597.
- Tokumitsu M, Kadohama T, Ogawa K (1994) Infrequent loss of heterozygosity and mutation of the p53 gene in immortal and transformed mouse embryo fibroblasts. Mol Carcinog 10: 52-57. [Crossref]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW (1992) A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol 12: 2866-2871. [Crossref]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, et al. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817-825. [Crossref]
- Chen CY, Oliner JD, Zhan Q, Fornace AJ Jr, Vogelstein B, et al. (1994) Interactions between p53 and MDM2 in a mammalian cell cycle checkpoint pathway. Proc Natl Acad Sci U S A 91: 2684-2688. [Crossref]
- Zauberman A, Barak Y, Ragimov N, Levy N, Oren M (1993) Sequence-specific DNA binding by p53: identification of target sites and lack of binding to p53 - MDM2 complexes. EMBO J 12: 2799-2808. [Crossref]
- Nozaki T, Masutani M, Akagawa T, Sugimura T, Esumi H (1994) Suppression of G1 arrest and enhancement of G2 arrest by inhibitors of poly(ADP-ribose) polymerase: possible involvement of poly(ADP-ribosyl)ation in cell cycle arrest following gamma-irradiation. Jpn J Cancer Res 85: 1094-1098. [Crossref]
- Fuchs SY, Adler V, Buschmann T, Wu X, Ronai Z (1998) Mdm2 association with p53 targets its ubiquitination. Oncogene 17: 2543-2547. [Crossref]
- Juven T, Barak Y, Zauberman A, George DL, Oren M (1993) Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene 8: 3411-3416.
- Lu X, Lane DP (1993) Differential induction of transcriptionally active p53 following UV or ionizing radiation: defects in chromosome instability syndromes? Cell 75: 765-778.
- Wieler S, Gagné JP, Vaziri H, Poirier GG, Benchimol S (2003) Poly(ADP-ribose) polymerase-1 is a positive regulator of the p53-mediated G1 arrest response following ionizing radiation. J Biol Chem 278: 18914-18921.




