MicroRNA’s (miRNA’s) are non-coding RNA’s that can regulate gene expression. miRNA’s are small (~22 nt) and can bind by complementation to mRNA’s resulting in direct degradation and/or interference with translation. We used paired-end RNAseq and small RNAseq to examine expression of mRNA’s and miRNA’s, respectively, in total RNA’s isolated from frontal cortex of Alzheimer’s disease (AD) and control (CTL) subjects. mRNA’s were aligned to the hg38 human genome using Tophat2/Bowtie2 and relative expression levels determined by Cufflinks as FPKM (fragments per kilobase of exon per million mapped reads). miRNA’s were aligned to the hg19 human genome and quantitated as number of mature known miRNA reads with miRDeep*. Using a false-discovery rate (FDR) of 10% (FDR<0.10) applied to 11,794 genes with FPKM > 2.0, we identified 55 genes upregulated in AD and 191 genes downregulated in AD. The top 145 miRNA’s with the highest mean expression were mostly under-expressed in AD (132/145 miRNAs had AD/CTL < 1.0)). AD samples had increased coefficients of variation for miRNA and mRNA expression, implying greater heterogeneity. miRTAR analysis showed that 32 (58%) of the mRNA genes upregulated in AD could be controlled by one or more of 60 miRNA’s under-expressed >1.5-fold in AD (AD/CTL < 0.67). AD frontal cortex gene expression analysis showed extensive regulation by miRNAs that appear to interact with the majority of overexpressed protein coding genes. The temporal sequence of appearance in AD brain and etiologies of this extensive, heterogeneous regulation by miRNA’s are unknown. (243 words)
RNA-sequencing, miRNA, frontal cortex, gene expression
Alzheimer’s disease (AD) most often occurs sporadically (>95%) and is the leading cause of dementia, which, according to Alzheimer’s Disease International (World Alzheimer Report 2010, “The Global Economic Impact of Dementia”), results in costs that represent the world’s 18th largest economy and is highest per capita in North American countries. It is estimated that the United States currently spends ~220 billion dollars per year for dementia care, most of which is due to AD.
AD progresses clinically through loss of one or more domains of cognition, known as mild cognitive impairment (MCI) [1-3]. Individuals with MCI have ~50% chance of progressing into AD dementia over 3-5 years and demonstrate impaired cerebral cortical glucose metabolism [3-11] and increased oxidative stress damage products [12]. These findings suggest an early impairment of bioenergetic function potentially arising from oxidative stress damage.
Although advanced AD brains demonstrate accumulation of beta amyloid peptide in extracellular “plaques”, and hyper-phosphorylated intraneuronal tau seen as “neurofibrillary tangles”, the hypotheses advancing causality of either of these proteins in soluble or aggregated forms for genesis or progression of AD dementia remain controversial [13]. Strategies of reducing beta amyloid protein levels, either by antibody clearance or synthesis inhibition, have to date not altered dementia progression [14-19]. These results have stimulated some to question the “amyloid cascade hypothesis” of AD pathogenesis, but the interpretations of these negative clinical research results remain controversial [13,18,19].
Improvements in sequencing technologies have allowed development of “next-generation sequencing” (NGS), which in an unbiased manner sequences fragments of DNA or cDNA’s. Alignment algorithms available in the public domain align these fragments to genomes, allowing quantitation of coding gene cDNA abundance, gene mutations, exon-intron boundaries, among many possibilities. The more recent development of technologies that allow small segment sequencing has allowed exploration of non-coding RNA’s, that include micro RNA’s (miRNA’s), small RNA’s whose genes exist on the intergenic DNA formerly referred to as “junk DNA”, but now recognized to contain coding sequences for a variety of small RNA’s whose functions are being explored.
miRNA’s are now considered to be part of the gene expression regulatory system, along with histone post-translational modifications and DNA methylation. miRNA’s, of which there are thousands currently known in the human genome, promiscuously regulate expression of many genes. A given miRNA can control expression of dozens of mRNA’s, by both binding tightly to the untranslated regions (UTR’s) of a gene, inducing degradation of that mRNA, or by interfering with the translation of the mRNA, reducing the resulting protein content. Likewise, a given mRNA can have many miRNA’s that regulate it with varying degrees of specificity.
Thus, modern NGS studies of gene expression can now include identification and quantitation of miRNA’s as regulators of gene expression. With this approach, a more comprehensive picture of how gene expression changes are regulated can emerge.
Brain samples
Brain samples were collected over multiple years by the University of Virginia Brain Resource Facility. Individuals, who died with ante-mortem diagnoses of Alzheimer’s dementia, after provision of informed consent by next of kin, underwent autopsy with brain removal. The brain was divided sagittally and half was immersed in formalin for histopathological diagnosis. The remaining half was sectioned coronally. From these sections, representative areas were dissected and rapidly frozen in liquid nitrogen-chilled 2-methlybutane. Both flash frozen blocks and coronal sections were stored at -80 degrees. For the present study, we restricted ourselves to samples that were autopsied and frozen within 12 hours of death. All brains were examined by a neuropathologist to confirm diagnosis of AD (or CTL).
Total RNA isolation
For this study, 25, 20 um cryostat sections of approximately 1 by 1.5 cm blocks of frontal cortex grey matter were collected and immediately placed in Qiazol. Total RNA was isolated according to Qiagen miRNeasy instructions with sonication of the tissue in Qiazol, on-column DNase digestion and an additional RPE buffer wash. RNA quality was assessed on an Agilent BioanalyzerÒ capillary gel electrophoresis system, and RNA was quantitated using a Nanodrop spectrophotometer. Total RNA samples were sent for RNA sequencing (CoFactor Genomics). CoFactor carried out the library preparation, quantitation and Illumina sequencing (see below for details).
RNA sequencing
Ribo-depletion, NGS library preparation: Total RNA isolated by us was processed for library construction by Cofactor Genomics (http://cofactorgenomics.com, St. Louis, MO) according to the following procedure: Briefly, species specific rRNA-probes were hybridized to total RNA in order to remove nuclear-encoded and mitochondrial contaminating ribosomal RNA from the sample. The resulting ribo-depleted RNA was then fragmented. First-strand cDNA synthesis was performed using reverse transcriptase and random primers in the presence of Actinomycin D, followed by second-strand cDNA synthesis with DNA polymerase I and RNase H. Double-stranded cDNA was end-repaired and A-tailed for subsequent adaptor ligation. Indexed adaptors were ligated to the A-tailed cDNA. Enrichment by PCR was performed to generate the final cDNA sequencing library. Libraries were sequenced as paired-end 150 base pair reads on a NextSeq500 (Illumina, San Diego, CA) following the manufacturer's protocols.
Small RNA size selection, NGS library preparation: Total RNA was processed for library construction by Cofactor Genomics (http://cofactorgenomics.com, St. Louis, MO) according to the following procedure: Briefly, adapters were ligated to the 5’ and 3’ ends of the RNA, allowing cDNA to be generated with a reverse transcriptase. Following a dual-SPRI size selection with Ampure XP beads, PCR was used to add indexes and amplify adapter-ligated fragments. A final size selection was performed using a Pippin HT instrument (Sage Science, Beverly, MA) to enrich for miRNA inserts. Libraries were sequenced as single-end 75 base pair reads on a NextSeq500 (Illumina, San Diego, CA) following the manufacturer’s protocols.
Paired-end mRNA libraries
We utilized a format similar to that described in our earlier work [20]. Briefly, compressed, paired-end sequencing files in FastQ format for each mRNA sample were downloaded, decompressed and examined in FastQC. They were then trimmed of Illumina sequencing adapters using Trimmomatic with Phred scores of 20 (99% minimum sequencing accuracy) and re-examined in FastQC to be certain all adapters had been removed. The trimmed sequences were aligned against the hg38 version of the human genome using Tophat2/Bowtie2, then quantitated against the hg38 human transcriptome using Cufflinks. Mean FPKM (fragments per kilobase of exon per million reads) were used for estimates of relative gene expression.
Single-end sequencing files were aligned against the hg19 version of the human genome using miRDeep* [21]. Total reads for known miRNA’s were manually aligned for each CTL and AD sample. miRNA target prediction of the miRNA expression data was carried out using miRTAR [22] with the default thermodynamic settings for miRNA-mRNA interactions. The gene lists studied included all miRNA’s where the mean AD/mean CTL ratio was <0.67 (ie, the miRNA’s were under-expressed in AD samples > 1.5-fold), and the mRNA genes were significantly (FDR < 10%) overexpressed. miRTAR identified 54 (58%) of the significantly overexpressed mRNA’s in AD samples that could be regulated by 16 miRNA’s underexpressed in AD brain.
qPCR
Primers for qPCR assay of gene expression were designed using BLAST Primer Design
(https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Primer pairs (n=12) for qbasePLUS/GeNormÒ determination of the optimum reference (“housekeeping”) genes to use for data normalization were obtained from Primer Design (http://www.primerdesign.co.uk/products/9460-genorm-kits) and used according to manufacturer instructions. qPCR was carried out in 96-well PCR plates using a BioRad CFX thermocycler and SybrGreen detection of DNA pairs. The GeNorm algorithm in qbasePLUS (v 3.1) was used to determine the least variable reference genes (CYC1, ATP5A1, ACTB) used to normalize gene expression results in qbasePLUS.
Statistics
All statistical processing used Prism v 7.0a (GraphPad) mRNA gene expressions were analyzed by multiple t-tests with two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli to determine False Discovery Rates (FDR) [23]. We report genes that were expressed with FDR < 10% performed by Prism software. Otherwise, 2-way ANOVA’s were used to determine significance (P<0.05).
Supplemental Table 1 shows all FPKM values for mRNA’s identified in frontal cortex post-mortem samples by paired-end RNA-seq from Tophat2/Bowtie2 alignment against the hg38 human genome and Cufflinks quantitation against the hg38 human transcriptome, for coding genes with mean FPKM>2.0, after removal of duplicate samples (n=11,795 total). We studied RNA’s isolated from frontal cortex samples harvested from 10 persons with AD and 9 CTL individuals, all collected under informed consent and examined by a neuropathologist. We used the Qiagen miRNeasyÒ system to isolate total RNA, which includes small RNA’s. Table 1 lists the demographics on the brain samples and their quality assayed with the Agilent BioanalyzerÒ system. There was no difference between the two groups (AD, CTL) in terms of mean RNA quality numbers. (4.7) The AD population was slightly older (but not significantly so) at death (AD = 78 +/- 3 years; CTL = 71 +/- 4 years; p = 0.19 by t-test).
Table 1. Age at Death and RNA Quality (RIN) for Postmortem Frontal Cortex Samples
Age at death AD |
Age at death CTL |
RIN AD |
RIN CTL |
92 |
63 |
5.1 |
5 |
66 |
83 |
5 |
4.9 |
81 |
53 |
4.8 |
4.9 |
73 |
88 |
4.7 |
4.7 |
69 |
87 |
4.4 |
4.6 |
90 |
70 |
4.3 |
4 |
78 |
71 |
4 |
4 |
77 |
53 |
4 |
5.3 |
78 |
71 |
4 |
|
77 |
|
6.9 |
|
78 +/- 3 |
71 +/- 4 |
4.7 +/- 0.3 |
4.7 +/- 0.1 |
p=0.19 |
|
p=0.97 |
|
Coding gene RNAseq was carried out as described in our earlier work [20], which provides links to all software algorithms and Unix texts used. Briefly, total RNA samples were treated to remove ribosomal RNA’s (Ribo-FreeÒ, Illumina) then Illumina multiplex sequencing oligonucleotides were attached. The libraries were paired-end sequenced on an Illumina (San Diego, CA) NextSeq 500 at ~ 60 million base depths. The resulting files in FastQ format were decompressed and examined with FastQC before and after trimming, which was carried out with Trimmomatic and minimum Phred scores of 20 (99% sequencing accuracy). Subsequent bioinformatics pipeline involved alignment against the hg38 version of the human genome with Tophat2/Bowtie2 and quantitation of expression against the hg38 human transcriptome using Cufflinks.
Table 2 lists the genes identified with FDR (q values) of 10% or less as over-expressed or under-expressed in the AD brain samples, along with their AD/CTL expression ratios determined from the mean FPKM values.
Table 2. Genes Over-expressed and Under-expressed in AD Frontal Cortex
gene |
q value |
AD/CTL |
gene |
q value |
AD/CTL |
HSPB1 |
0.09 |
3.71 |
BRK1 |
0.09 |
0.87 |
RBM14 |
0.09 |
2.84 |
KLHL28 |
0.09 |
0.87 |
RSAD1 |
0.09 |
2.82 |
FAF2 |
0.07 |
0.86 |
GSN |
0.09 |
2.79 |
USP10 |
0.07 |
0.86 |
PAXBP1 |
0.1 |
2.74 |
RPRD1B |
0.09 |
0.86 |
SPR |
0.09 |
2.06 |
KIAA0368 |
0.08 |
0.86 |
RBM4 |
0.07 |
2.04 |
GABARAPL2 |
0.09 |
0.86 |
CCNG1 |
0.07 |
2.04 |
TMED2 |
0.1 |
0.86 |
BST2 |
0.09 |
1.98 |
BCL2L2 |
0.08 |
0.85 |
TNFRSF25 |
0.1 |
1.92 |
LARP4B |
0.08 |
0.85 |
ZNF358 |
0.09 |
1.91 |
IARS2 |
0.1 |
0.85 |
TREM2 |
0.07 |
1.91 |
KIAA1468 |
0.09 |
0.85 |
LAMB2 |
0.09 |
1.9 |
TMEM30A |
0.1 |
0.85 |
ZDHHC11 |
0.07 |
1.89 |
THRAP3 |
0.09 |
0.85 |
ZSWIM8 |
0.09 |
1.88 |
CACUL1 |
0.09 |
0.85 |
INPP5B |
0.07 |
1.86 |
FAM98B |
0.09 |
0.85 |
DYRK1A |
0.08 |
1.85 |
IREB2 |
0.09 |
0.84 |
KIAA0930 |
0.09 |
1.82 |
SENP5 |
0.07 |
0.84 |
ITGAV |
0.08 |
1.77 |
HEATR5B |
0.07 |
0.84 |
LZTR1 |
0.09 |
1.75 |
ATR |
0.09 |
0.84 |
TKT |
0.09 |
1.72 |
CRBN |
0.07 |
0.84 |
GSTP1 |
0.07 |
1.69 |
WBP4 |
0.1 |
0.84 |
C9orf114 |
0.07 |
1.68 |
SNRPE |
0.09 |
0.84 |
MAP3K3 |
0.08 |
1.66 |
FAM199X |
0.07 |
0.84 |
PLXNB1 |
0.09 |
1.62 |
MRPS16 |
0.07 |
0.84 |
PLXND1 |
0.09 |
1.61 |
EXOC4 |
0.07 |
0.84 |
KLHL21 |
0.08 |
1.6 |
VAPA |
0.07 |
0.83 |
FLCN |
0.07 |
1.54 |
SEC63 |
0.08 |
0.83 |
KAT2A |
0.09 |
1.52 |
SNX13 |
0.1 |
0.83 |
INPPL1 |
0.07 |
1.51 |
DNAJC18 |
0.09 |
0.83 |
RPAP1 |
0.09 |
1.51 |
KLHDC10 |
0.08 |
0.83 |
MICAL3 |
0.09 |
1.5 |
TMEM50A |
0.09 |
0.83 |
TYK2 |
0.09 |
1.48 |
NLGN1 |
0.07 |
0.83 |
H2AFX |
0.08 |
1.47 |
HNRNPA0 |
0.07 |
0.83 |
ACTN4 |
0.09 |
1.46 |
TBC1D19 |
0.09 |
0.83 |
OGDH |
0.07 |
1.46 |
C4orf3 |
0.07 |
0.83 |
ANKS6 |
0.07 |
1.45 |
FBXW2 |
0.07 |
0.83 |
STARD10 |
0.09 |
1.43 |
FAM192A |
0.07 |
0.82 |
SMG9 |
0.07 |
1.42 |
NRIP1 |
0.09 |
0.82 |
GEMIN4 |
0.08 |
1.38 |
POLE3 |
0.07 |
0.82 |
WASH5P |
0.09 |
1.38 |
CDKL2 |
0.09 |
0.82 |
SMG5 |
0.09 |
1.37 |
MRPL47 |
0.08 |
0.82 |
TMEM109 |
0.08 |
1.36 |
VMP1 |
0.09 |
0.82 |
SNORA37 |
0.09 |
1.35 |
SUDS3 |
0.07 |
0.82 |
SNORA73B |
0.09 |
1.35 |
NMD3 |
0.09 |
0.82 |
MCF2L |
0.1 |
1.35 |
OSBP |
0.09 |
0.82 |
GCN1 |
0.08 |
1.34 |
MANSC1 |
0.09 |
0.82 |
KANTR |
0.08 |
1.33 |
MFN1 |
0.08 |
0.81 |
FKBP4 |
0.08 |
1.32 |
KIAA1715 |
0.09 |
0.81 |
FADD |
0.09 |
1.31 |
MATR3 |
0.07 |
0.81 |
SAT2 |
0.08 |
1.3 |
TRIM27 |
0.1 |
0.81 |
SNORA73A |
0.08 |
1.3 |
NALCN |
0.09 |
0.81 |
CEP250 |
0.08 |
1.29 |
SKP1 |
0.09 |
0.81 |
RILPL1 |
0.08 |
1.27 |
MRPS35 |
0.07 |
0.81 |
IFFO2 |
0.09 |
1.19 |
GTPBP4 |
0.08 |
0.81 |
VMA21 |
0.09 |
0.81 |
ISPD |
0.07 |
0.74 |
TBL1XR1 |
0.07 |
0.81 |
RABL3 |
0.07 |
0.74 |
NOA1 |
0.09 |
0.81 |
BZW2 |
0.07 |
0.74 |
TOP2B |
0.08 |
0.8 |
SLC30A9 |
0.08 |
0.74 |
SYT11 |
0.08 |
0.8 |
RPARP-AS1 |
0.1 |
0.74 |
KIAA1279 |
0.09 |
0.8 |
SLC35B3 |
0.1 |
0.74 |
RAB18 |
0.07 |
0.8 |
ALDH5A1 |
0.08 |
0.73 |
DHX36 |
0.07 |
0.8 |
TCEAL7 |
0.09 |
0.73 |
DNTTIP2 |
0.09 |
0.8 |
PLAA |
0.07 |
0.73 |
AKAP11 |
0.07 |
0.8 |
RPL23AP64 |
0.07 |
0.73 |
FBXL20 |
0.07 |
0.8 |
PTS |
0.09 |
0.73 |
RPAP3 |
0.09 |
0.8 |
TSNAX |
0.08 |
0.72 |
SAR1A |
0.07 |
0.8 |
CARF |
0.09 |
0.72 |
YME1L1 |
0.09 |
0.8 |
DIRC2 |
0.09 |
0.72 |
TAB3 |
0.09 |
0.8 |
SEC23IP |
0.09 |
0.72 |
ANKRD13C |
0.07 |
0.8 |
C5orf22 |
0.08 |
0.72 |
SEC62 |
0.07 |
0.79 |
TMEM150C |
0.08 |
0.72 |
HMG20A |
0.09 |
0.79 |
MPHOSPH6 |
0.07 |
0.71 |
GNPTAB |
0.07 |
0.79 |
ABCA11P |
0.07 |
0.71 |
PLRG1 |
0.08 |
0.79 |
TASP1 |
0.09 |
0.71 |
TSPYL5 |
0.1 |
0.79 |
ZNF669 |
0.09 |
0.71 |
CHP1 |
0.09 |
0.79 |
RASGRP1 |
0.09 |
0.71 |
TSN |
0.07 |
0.79 |
ZCCHC17 |
0.09 |
0.71 |
GGNBP2 |
0.07 |
0.79 |
ZNF136 |
0.09 |
0.71 |
LRPPRC |
0.08 |
0.79 |
HIGD1A |
0.09 |
0.71 |
C11orf58 |
0.08 |
0.79 |
IMPACT |
0.09 |
0.71 |
NOCT |
0.1 |
0.79 |
SNX4 |
0.09 |
0.7 |
PRKAA1 |
0.07 |
0.79 |
EXOSC9 |
0.09 |
0.7 |
GOLT1B |
0.08 |
0.79 |
TM6SF1 |
0.09 |
0.7 |
EAF1 |
0.09 |
0.79 |
LINC01250 |
0.09 |
0.7 |
PSME3 |
0.07 |
0.78 |
PEX7 |
0.09 |
0.69 |
AVL9 |
0.07 |
0.78 |
MIR4458HG |
0.07 |
0.69 |
RINT1 |
0.07 |
0.78 |
TAF9 |
0.07 |
0.68 |
MTMR7 |
0.07 |
0.78 |
ZNF449 |
0.09 |
0.68 |
WAC-AS1 |
0.07 |
0.78 |
SUB1 |
0.09 |
0.68 |
PTPMT1 |
0.09 |
0.78 |
ACADSB |
0.07 |
0.68 |
RFWD2 |
0.07 |
0.78 |
PTP4A1 |
0.07 |
0.67 |
ATXN10 |
0.07 |
0.78 |
RBM11 |
0.09 |
0.67 |
PSMD1 |
0.08 |
0.78 |
FBXO22 |
0.07 |
0.67 |
LINC00657 |
0.07 |
0.78 |
PPP1R3C |
0.08 |
0.67 |
IMPAD1 |
0.07 |
0.78 |
DHDDS |
0.09 |
0.67 |
AP3S2 |
0.07 |
0.78 |
LSM5 |
0.07 |
0.67 |
CYP2R1 |
0.09 |
0.78 |
PPP1R2 |
0.08 |
0.67 |
NRAS |
0.07 |
0.78 |
FOPNL |
0.07 |
0.66 |
GMFB |
0.07 |
0.78 |
ZNF441 |
0.07 |
0.66 |
POLR3F |
0.09 |
0.78 |
TATDN1 |
0.07 |
0.65 |
NAV3 |
0.09 |
0.77 |
HS3ST2 |
0.08 |
0.65 |
CRLS1 |
0.07 |
0.77 |
TARBP1 |
0.09 |
0.65 |
ZNF25 |
0.1 |
0.77 |
STX16- NPEPL1 |
0.09 |
0.65 |
MARCKS |
0.09 |
0.77 |
SAYSD1 |
0.08 |
0.64 |
SRSF8 |
0.09 |
0.77 |
MGAT4C |
0.08 |
0.64 |
RAB2A |
0.09 |
0.77 |
LOC728730 |
0.07 |
0.64 |
TIPRL |
0.09 |
0.77 |
NT5DC3 |
0.08 |
0.64 |
SPCS1 |
0.09 |
0.77 |
ENKUR |
0.09 |
0.63 |
RIOK3 |
0.09 |
0.77 |
ZRANB3 |
0.08 |
0.62 |
STX12 |
0.07 |
0.77 |
SLC8A3 |
0.1 |
0.6 |
PRPF4 |
0.09 |
0.76 |
C2orf80 |
0.07 |
0.57 |
PWP1 |
0.1 |
0.76 |
MIR22HG |
0.09 |
0.56 |
TMEM41A |
0.07 |
0.76 |
KIAA1524 |
0.08 |
0.56 |
USP14 |
0.07 |
0.76 |
TRABD2A |
0.07 |
0.56 |
SMIM10L1 |
0.07 |
0.75 |
SCARNA9L |
0.09 |
0.54 |
SHFM1 |
0.07 |
0.75 |
LINC00936 |
0.1 |
0.52 |
EIF2S1 |
0.07 |
0.75 |
ZDHHC16 |
0.1 |
0.5 |
WBP11 |
0.07 |
0.75 |
ADTRP |
0.07 |
0.41 |
ALG14 |
0.09 |
0.75 |
TMEFF1 |
0.07 |
0.41 |
WDR17 |
0.09 |
0.75 |
DPH2 |
0.07 |
0.38 |
ZMAT2 |
0.08 |
0.75 |
GNG4 |
0.09 |
0.37 |
SLC25A16 |
0.07 |
0.74 |
CYP26B1 |
0.07 |
0.32 |
Gene expression was assayed by RNA-seq as described in Methods. After removal of duplicates, all AD and CTL frontal cortex genes with mean FPKM > 2.0 (n=11,795) were subjected to multiple t-test comparison with FDR correction < 10% (q < 0.10) applied for multiple comparisons.
To create Table 3, we used qPCR to assay all AD and CTL cDNA’s for the 4 genes yielding the highest AD/CTL ratios (HSPB1, RBM14, RSAD1, GSN) and the 4 genes yielding the lowest AD/CTL ratios (TMEFF1, DPH2, GNG4, CYP26B1) (see Table 2). After normalizing each sample to the geometric mean of the 3 reference genes found to be least variable by GeNorm, we found with qbasePLUS (Table 3) that 7 out of 8 genes had expression levels qualitatively similar (increased or decreased in AD) by qPCR compared to RNA-seq. The one exception was CYP26B1 that was slightly overexpressed in AD by qPCR but underexpressed by RNA-seq.
Table 3. Comparison of mean AD/CTL gene expression ratios between qPCR and RNA-seq
Gene |
AD/CTL (qPCR) |
AD/CTL (RNA-seq) |
CYP26B1 |
1.28 |
0.32 |
GNG4 |
0.72 |
0.37 |
DHP2 |
0.73 |
0.38 |
TMEFF1 |
0.46 |
0.41 |
GSN |
2 |
2.79 |
RSAD1 |
1.42 |
2.82 |
RMB14 |
1.47 |
2.84 |
HSPB1 |
2.53 |
3.71 |
Small RNA sequencing was carried out on each total RNA sample that was also sequenced for mRNA genes as above. Sequencing files were first examined for residual sequencing library adapters and then restricted with Trimmomatic to Phred scores of 30 (sequencing accuracy > 99.9%), a “sliding window” of 4 bases, each requiring a minimum Phred score of 30, and a maximum length of 24 bases. In this manner, we wished to restrict the small miRNA’s identified to those of an appropriate size and very high sequencing accuracy.
The resulting files were processed individually using miRDeep* [21] alignment (operating in JavaÒ mode) against the hg19 human genome. Known miRNA’s from each of the CTL and AD samples were manually curated to align expression (as number of reads of each mature miRNA) across both groups. From this Table (Supplemental Table 2) the mean, SD, coefficient of variance (CV) and mean AD/mean CTL ratios were calculated. The minimum number of samples for which a miRNA was present was set to be 6 for the CTL samples and 7 for the AD samples. In Supplemental Table 2, the mean number of samples for detected miRNA’s was 8.7 for CTL and 9.5 for AD.
Figure 1 shows the results of miRNA quantitation, with results expressed as % of mean CTL values +/- SEM. 132 out of 145 miRNA’s were under-expressed in AD samples. (AD/CTL < 100% mean CTL)
Expression (+/- SEM) of Top 145 miRNA’s in Ftl Ctx p=0.0323 CTL vs AD 2-way AN
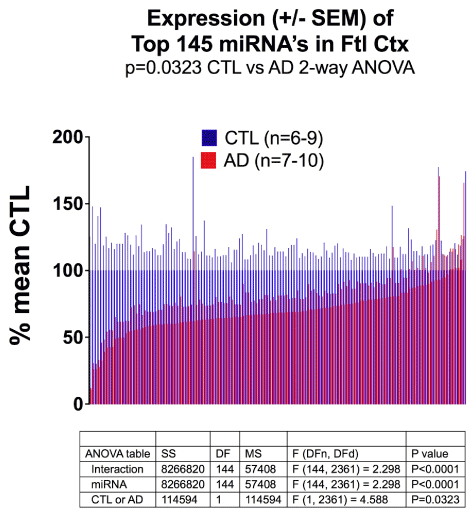
Figure 1. MicroRNA Expression is reduced in AD Frontal Cortex. Shown are miRNA expression results (as number of mature reads), normalized to mean CTL values, plotted as average % mean CTL +/- SEM. The results for AD (red) were reduced in 132/145 miRNA’s. See Methods for details.
miRTAR [22] was then used to assign interactions of each miRNA most under-expressed in the AD samples (mean AD/mean CTL < 0.67) with the mRNA genes over-expressed in the AD samples with FDR<10%. Default miRTAR free energy values of miRNA binding were used, and interactions were sought with only the 3’ UTR of each gene.
Table 4 shows the results of the miRTAR analysis of the interactions among miRNA’s (under-expressed in AD) and mRNA’s (over-expressed in AD) identified in the RNA seq studies. We identified 32 mRNA’s (out of 55 over-expressed in AD, 58%) that could be regulated by 60 miRNA’s underexpressed in AD, using the conservative thermodynamic default values of miRNA-mRNA interactions in miRTAR.
Table 4. miRTAR Analysis of Genes Potentially Downregulated by miRNA’s
miRNA |
Gene(s) potentially downregulated (3’ UTR) |
single miRNA: single mRNA |
|
a. hsa-mir-1250 |
MAP3K3 |
b. hsa-mir-126 |
GEMIN4 |
c. hsa-mir-161d |
CCNG1 |
d. hsa-mir-340 |
ITGAV |
single miRNA: two mRNA’s |
|
a. hsa-mir-136 |
FLCN, MICAL3 |
b. hsa-mir-148a |
DYRK1A, LAMB2 |
c. hsa-mir-212 |
PLXNB1, PLXND1 |
d. hsa-mir-222 |
OGDH, RIPL1 |
single miRNA: three mRNA’s |
|
a. hsa-mir-184 |
BST2, IFFO2, INPPL1 |
single miRNA: four mRNA’s |
|
a. hsa-mir-134 |
DYRK1A, FADD, LZTR1, TMEM109 |
b. hsa-mir-2110 |
FLCN, IFFO2, MAP3K3, TMEM109 |
single miRNA: five mRNA’s |
|
a. hsa-mir-22 |
ANK56, FLCN, KLHL21, 0GDH |
single miRNA: six mRNA’s |
|
a. hsa-mir-15a |
BST2, CEP250, MICAL3, PLXNB1, TMEM109, TNFRSF25 |
single miRNA: seven mRNA’s |
|
a. hsa-mir-197 |
ANK56, DYRK1A, FLCN, INPP5B, ITGAV, PLXNB1, RBM4 |
single miRNA: nine mRNA’s |
|
a. hsa-mir-1301 |
CEP250, H2AFX, LAMB2, MAP3K3, MICAL3, SMG5, TMEM109, TYK2, ZDHHC11 |
single miRNA: thirteen mRNA’s |
|
a. has-mir-214 |
ACTN4, ANK56, GEMIN4, HSPB1, IFFO2, KLHL21, MICAL3, PLXNB1, PLXND1, SMG5, SPR, TMEM109 |
All miRNA’s were determined to be under-expressed in AD frontal cortex by miRDeep* and had mean AD/mean CTL ratios of <0.67 (>1.5-fold under-expression in AD). mRNA genes were determined by FDR<10% to be over-expressed in AD frontal cortex. miRTAR default values (MFE < -14 kcal/mol; Score > 140) was used to search for interactions among these miRNA’s and mRNA’s.
Of the 32 mRNA’s whose expressions were potentially regulated by miRNA’s as identified by miRTAR, we found that 7 (BST2, CEP250, GEMIN4, ITGAV, KLH21, LAMB2, SMG5) could be regulated by two miRNA’s; 3 (ANKS6, DYRK1A, MAP3K3) could be regulated by 3 miRNA’s; 4 (FLCN, IFFO2, MICAL3, PLXNB1) could be regulated by 4 miRNA’s; and 1 (TMEM109) could be regulated by 5 miRNA’s. We consider these 15 mRNA genes to be the most regulated by the miRNA’s we could identify in our AD brain samples.
Figure 2 shows the differences in coefficients of variation (CV) as a function of mean expression levels for the mRNA (bottom) and miRNA (top) genes identified in the AD and CTL populations. There was no apparent relationship among CV’s and mRNA expression levels. The CV’s of the AD samples for both mRNA and miRNA were significantly elevated compared to CTL, suggesting greater heterogeneity in the AD samples.
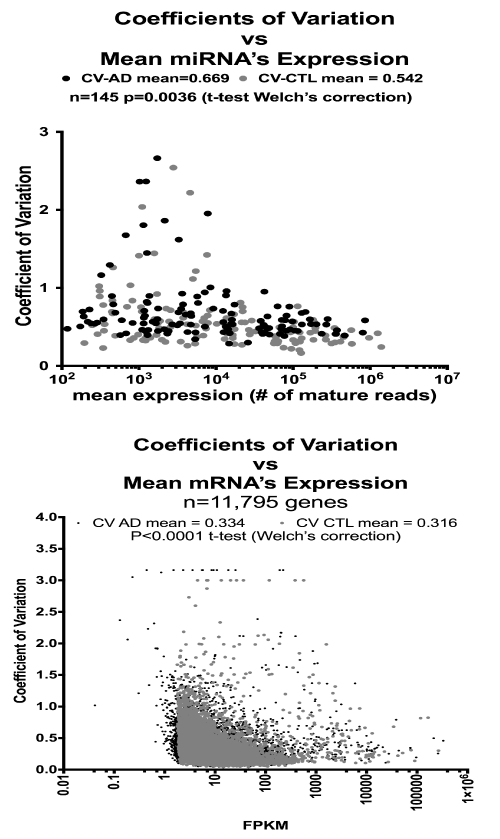
Figure 2. Coefficients of Variation are increased in AD miRNA’s and mRNA’s. Shown are individual mean values for coefficients of variation (CV; [standard deviation/mean]) on the Y-axes plotted against mean expression levels (logarithmic scale) on the X-axes. Results for miRNA’s (top graph) and mRNA’s (bottom graph) both reveal that the CV’s for the AD samples were greater than for the CTL samples. See Methods for details.
There are at least three major findings of our study:
First, we carried out RNA-seq of both coding genes and regulatory small RNA’s (miRNA’s) on total RNA’s isolated from frontal cortex brain samples of an AD cohort (n=10) and a CTL, disease-free, cohort (n=9). Using false-discovery rate correction for multiple samples (FDR < 10%), we found significant up-regulation of 55 genes and down-regulation of 191 genes in the AD population. Using DAVID analyses of the significantly over- and under-expressed genes in our AD samples (not shown), we could not determine any gene ontology groups or pathways that were significantly represented (Benjamini-corrected p values < 0.05). This speaks to the widespread heterogeneity of molecular genetic changes observed in our AD samples.
Second, our miRNA analysis with miRTAR suggests that ~58% of the AD upregulated genes could be regulated by miRNA’s that were under-expressed <1.5-fold in the AD samples. This result implies that miRNA regulation potentially plays a significant role in gene expression changes in AD. Because miRNA’s can regulate multiple coding genes at both the mRNA and protein translation levels, AD brain, at least at the advanced stages, appears to experience multiple and wide-ranging molecular genetic abnormalities likely derived from alterations in miRNA regulation.
The third major finding is the increased heterogeneity (AD>CTL) in the brain samples for expression of both mRNA’s and miRNA’s. This result suggests that no single molecular cause for AD is likely to be uncovered, and that multiple molecular genetic abnormalities may exist among individuals that result in shared clinical phenotypes over time. Identifying molecular genetic subgroups may allow protective therapies to be “personalized”.
There are several important caveats about our study that potentially limit interpretation:
First, we were restricted to use of post-mortem tissues from persons who died with advanced disease. Our findings may represent “survival” strategies of neurons and glia in these samples. One cannot assume that we have identified pathogenic patterns of gene expression or miRNA depletion.
Second, we studied a limited number of samples. The current high costs of RNA-seq limits studies in terms of the total population size examined. Since AD afflicts > 5 million persons in the US alone, extrapolation of our findings from a limited cohort to this very large population is risky.
Third, our RNA quality was not ideal, compared to total RNA quality isolated from quickly processed human biopsies, animal tissues or cells. This is an inevitable limitation of post-mortem human brain samples; although some improvements in RNA quality have been reported with “rapid” autopsy/freezing procedures. Because our AD and CTL samples did not differ in overall RNA quality, it is unlikely that our results can be explained on the basis of variable RNA degradations. However, that consideration must be kept in mind.
Fourth, we cannot speculate as to the temporal sequence of appearance of the abnormalities we found. If these abnormalities appear early in clinical disease evolution, then the earliest cognitive changes may signal marked disruption in the AD brain of molecular genetic harmony. If true, this suggests that disease-altering treatments should begin very early, prior to clinical deficits. This would require some type of risk stratification, similar to current approaches to vascular disease and lowering of blood cholesterol levels, as an example.
An alternative explanation is that we are describing late-appearing events. In this case, stratification of therapies in different molecular subgroups may identify specific populations that experience greater or lesser response to a given therapy at the time of symptom onset. In this scenario, therapies might be “personalized” based on molecular genetic profiles. It presently is unclear if this approach is even possible, much less meaningful to alter disease-modifying treatments of AD that to date have not succeeded. However, the degree of increased molecular genetic heterogeneity we observed in the AD population supports exploration of subpopulations in the AD community that could form the basis of stratifying future clinical trials.
Fifth, we did not carry out any miRNA expression studies in cell models to seek confirmation of mRNA changes. It would appear meaningful to overexpress the miRNA’s that modulated expression of multiple mRNA’s. As an example, five miRNA’s (mir-134 (AD/CTL = 0.541), mir-2110 (AD/CTL=0.557), mir-15a (AD/CTL=0.604), mir-1301 (AD/CTL=0.666), and mir-214 (AD/CTL=0.599)) are predicted by miRTAR to have significant thermodynamic interactions with TMEM109 mRNA. TMEM109, a gene coding for a transmembrane protein felt to yield a voltage-dependent ion channel and to mediate UV light-induced DNA damage, is one of the genes significantly (FDR<10%) overexpressed in our AD frontal cortex samples, consistent with the 5 indicated miRNA’s all being underexpressed in our AD brain samples.
Sixth, we used total tissue homogenates for RNA isolation. As a result, our findings represent a combination of RNA’s from multiple cell types, such as neurons (5-10% of cells), astrocytes (~80-85% of cells) and unknown but minor amounts of microglia, oligodendroglia and endothelial cells. We cannot assign the changes we observed to any particular cell type. This would require microdissection of specific cell types followed by RNA-seq.
There are multiple miRNA studies of AD brain tissues that use different assay methods, such as microarrays, qPCR and small RNA-seq (for a recent review, see [24]). Because miRNA’s are promiscuous in terms of the mRNA genes they can regulate, the varieties of AD tissues that have been examined, and the varying techniques to quantitate miRNA’s (ie, qPCR, microarray, small RNA-seq), there is presently no “consensus” as to the deregulation of which miRNA’s are critical to AD pathophysiology [24]. Our approach, combining traditional RNA-seq of gene cDNA’s and non-coding small RNA-seq from the same total RNA samples, combined with predictive bioinformatics tools, yields interactions among mRNA’s and miRNA’s that may be relevant to AD brain.
A particular miRNA consistently felt to be critical to AD pathogenesis is miR-132 [25], which has been found to be extensively down-regulated. We also found expression of miR-132 to be down-regulated in our samples, with an AD/CTL ratio of 0.389. (miR-132 is highlighted in Supplemental Table 2).
The nature of our study is primarily descriptive rather than mechanistic. For instance, we showed (see above) that multiple miRNA’s we found were under-expressed in AD cortex samples (thus relatively over-expressed in CTL samples), were predicted thermodynamically to interact with a single mRNA (TMEM109). Thus any or all of these miRNA’s could bind to and lead to the degradation of TMEM109 mRNA in CTL brain samples. This would constitute a pre-translational regulation of gene expression, but it does not inform us as to possible translational mechanisms of controlling TMEM109 protein levels that these miRNA’s are also capable of.
2021 Copyright OAT. All rights reserv
Our study has multiple caveats and limitations (discussed above), but there are several predictions that are experimentally testable in human iPSC-derived neural cells. Testing these predictions would be important to define mechanistic interpretations of our findings relevant to AD pathogenesis. One prediction would be that expression of genes both overexpressed in AD brain and potentially regulated by several miRNA’s underexpressed in AD brain (such as TMEM109 that is potentially regulated by 5 miRNA’s) would adversely affect iPSC-derived neural stem cells derived from disease-free (CTL) persons. We are presently carrying out that experiment. Other genes (overexpressed-in-AD brains) potentially regulated by multiple miRNA’s could be tested singly or in combination. Should these overexpressions reveal quantitative effects, conversion to a high-throughput format would allow screening of potential disease-altering medications.
Our findings support the concept that AD arises from dysfunction of multiple molecular systems and is heterogeneous across individuals in terms of which molecular systems have greater or lesser degrees of dysfunction. A therapeutic strategy for mild cognitive impairment and early AD has been devised, which attempts to improve simultaneously multiple nutritional, endocrinological and mitochondrial dysfunctions. The MEND protocol (Metabolic Enhancement for Neuro Degeneration) has achieved some success in subjects with early dementia [26]. One potential refinement of such a protocol would be to examine how the MEND approach alters gene and miRNA expression in peripheral cells, or perhaps neural membrane exosomes [27].
In summary, we found using RNA-seq and small RNA-seq on the same RNA samples that in AD frontal cortex expression of multiple genes (mRNA) was significantly up or down-regulated. ~58% of the up regulated genes could derive from effects of miRNA’s that were under-expressed in AD brains. Because we did not examine the results of overexpressing individual miRNA’s in cells, we can provide only correlational changes and cannot speak to potential causation of the miRNA changes we observed.
We hope to stimulate interest in the problems of molecular heterogeneity in a complex brain disease and therapeutic stratification for future clinical studies. In addition, if AD (at least, AD at end stage, which is what our post-mortem samples represent) includes significant miRNA control of gene expression, what factors mediate those changes? Is miRNA expression repressed in AD brain in response to “environmental” changes or other stresses? That such regulations appear to be present in AD support the hypothesis that variables of living can affect AD risk heterogeneously across individuals and that sporadically occurring AD should no longer be regarded as a monolithic illness with a common origin (or a common treatment).
Finally, our approach demonstrated the advantage of performing RNAseq for both mRNA’s and miRNA’s on the same RNA samples. By using this approach we were able to identify a core set of regulated mRNA’s in AD that may have their expression increased by loss of specific miRNA’s. Whether therapeutic strategies directed at multiple molecular systems, such as the MEND protocol that appears to improve cognitive function [26], alter expressions of miRNA’s or mRNA’s remains to be tested.
This research was supported by Neurodegeneration Therapeutics, Inc. (NTI) and the ALS Worldwide Foundation (ALSWW). Both NTI and ALSWW are 501(c)3 non-profit charitable research foundations. Neither sponsor had influence over the decision to publish or the timing of publication. We thank Dr. J. An for assistance with using miRDeep*.
Human subject protection and conflicts of interest
This study was conducted with attention to protection of human subjects. The human brain specimens used in this study were collected with informed consent of the families of the deceased. The samples are coded and identified only as age at death, gender, diagnoses, post-mortem intervals. JPB and PMK are officers of NTI but have no conflicts of interest with this study.
Author’s contributions
JPB and PMK designed the study. PMK maintained inventory of all brain samples, prepared brain sections, isolated and assayed RNA’s. JPB performed all bioinformatics assays and analyzed resulting data. JPB wrote the paper and both authors edited and agree with the final version. We thank Dr. Brent Wilson of CoFactor Genomics for assistance with RNAseq and small RNAseq studies and Dr. David G. Brohawn of Virginia Commonwealth University for early assistance with bioinformatic processing of RNAseq data.
- Cheng YW, Chen TF, Chiu MJ (2017) From mild cognitive impairment to subjective cognitive decline: conceptual and methodological evolution. Neuropsychiatr Dis Treat 13: 491-498. [Crossref]
- Gallagher D, Fischer CE, et al. (2017) Neuropsychiatric Symptoms in Mild Cognitive Impairment. Can J Psychiatry 62: 161-169. [Crossref]
- Sanchez-Catasus CA, Stormezand GN, van Laar PJ, De Deyn PP, Sanchez MA, et al. (2017) FDG-PET for Prediction of AD Dementia in Mild Cognitive Impairment. A Review of the State of the Art with Particular Emphasis on the Comparison with Other Neuroimaging Modalities (MRI and Perfusion SPECT). Curr Alzheimer Res 14: 127-142. [Crossref]
- Kemppainen N, Joutsa J, Johansson J, Scheinin NM, Nagren K, et al. (2015) Long-Term Interrelationship between Brain Metabolism and Amyloid Deposition in Mild Cognitive Impairment. J Alzheimers Dis 48: 123-133. [Crossref]
- Coutinho AM, Porto FH, Duran FL, Prando S, Ono CR, et al. (2015) Brain metabolism and cerebrospinal fluid biomarkers profile of non-amnestic mild cognitive impairment in comparison to amnestic mild cognitive impairment and normal older subjects. Alzheimers Res Ther 7: 58. [Crossref]
- Ito K, Fukuyama H, Senda M, Ishii K, Maeda K, et al. (2015) Prediction of Outcomes in Mild Cognitive Impairment by Using 18F-FDG-PET: A Multicenter Study. J Alzheimers Dis 45: 543-552. [Crossref]
- Bailly M, Destrieux C, Hommet C, Mondon K, Cottier JP, et al. (2015) Precuneus and Cingulate Cortex Atrophy and Hypometabolism in Patients with Alzheimer's Disease and Mild Cognitive Impairment: MRI and (18)F-FDG PET Quantitative Analysis Using FreeSurfer. Biomed Res Int 2015: 583931. [Crossref]
- Ota K, Oishi N, Ito K, Fukuyama H, Group S-JS, et al. (2016) Prediction of Alzheimer's Disease in Amnestic Mild Cognitive Impairment Subtypes: Stratification Based on Imaging Biomarkers. J Alzheimers Dis 52: 1385-1401. [Crossref]
- Xu L, Wu X, Li R, Chen K, Long Z, et al. (2016) Prediction of Progressive Mild Cognitive Impairment by Multi-Modal Neuroimaging Biomarkers. J Alzheimers Dis 51: 1045-1056. [Crossref]
- Caffarra P, Ghetti C, Ruffini L, Spallazzi M, Spotti A, et al. (2016) Brain Metabolism Correlates of the Free and Cued Selective Reminding Test in Mild Cognitive Impairment. J Alzheimers Dis 51: 27-31. [Crossref]
- Song IU, Choi EK, Oh JK, Chung YA, Chung SW (2016) Alteration patterns of brain glucose metabolism: comparisons of healthy controls, subjective memory impairment and mild cognitive impairment. Acta Radiol 57: 90-97. [Crossref]
- Pratico D, Clark CM, Liun F, Rokach J, Lee VY, et al. (2002) Increase of brain oxidative stress in mild cognitive impairment: a possible predictor of Alzheimer disease. Arch Neurol 59: 972-976. [Crossref]
- Smith MA, Drew KL, Nunomura A, Takeda A, Hirai K, et al. (2002) Amyloid-beta, tau alterations and mitochondrial dysfunction in Alzheimer disease: the chickens or the eggs? Neurochem Int 40: 527-531. [Crossref]
- Castellani RJ, Perry G (2012) Pathogenesis and disease-modifying therapy in Alzheimer's disease: the flat line of progress. Arch Med Res 43: 694-698. [Crossref]
- Panza F, Solfrizzi V, Imbimbo BP, Logroscino G (2014) Amyloid-directed monoclonal antibodies for the treatment of Alzheimer's disease: the point of no return? Expert Opin Biol Ther 14: 1465-1476. [Crossref]
- Salloway S, Sperling R, Fox NC, Blennow K, Klunk W, et al. (2014) Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer's disease. N Engl J Med 370, 322-333. [Crossref]
- Doody RS, Thomas RG, Farlow M, Iwatsubo T, Vellas B, et al. (2014) Phase 3 trials of solanezumab for mild-to-moderate Alzheimer's disease. N Engl J Med 370: 311-321. [Crossref]
- Karran E, De Strooper B (2016) The amyloid cascade hypothesis: are we poised for success or failure? J Neurochem 2: 237-252. [Crossref]
- Ricciarelli R, Fedele E (2017) The Amyloid Cascade Hypothesis in Alzheimer's Disease: It's Time to Change Our Mind. Curr Neuropharmacol 15: 926-935. [Crossref]
- Brohawn DG, O'Brien LC, Bennett JP, Jr. (2016) RNAseq Analyses Identify Tumor Necrosis Factor-Mediated Inflammation as a Major Abnormality in ALS Spinal Cord. PLoS One 11: e0160520. [Crossref]
- An J, Lai J, Lehman ML, Nelson CC (2013) miRDeep*: an integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res 41: 727-737. [Crossref]
- Hsu JB, Chiu CM, Hsu SD, Huang WY, Chien CH, et al. (2011) miRTar: an integrated system for identifying miRNA-target interactions in human. BMC Bioinformatics 12: 300. [Crossref]
- Benjamini Y, Yekutieli D (2005) Quantitative trait Loci analysis using the false discovery rate. Genetics 171: 783-790. [Crossref]
- Millan MJ (2017) Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer's disease: An integrative review. Prog Neurobiol 156: 1-68. [Crossref]
- Salta E, De Strooper B (2017) microRNA-132: a key noncoding RNA operating in the cellular phase of Alzheimer's disease. FASEB J 31: 424-433. [Crossref]
- Bredesen DE (2014) Reversal of cognitive decline: a novel therapeutic program. Aging (Albany NY) 6: 707-717. [Crossref]
- Mullins RJ, Mustapic M, Goetzl EJ, Kapogiannis D (2017) Exosomal biomarkers of brain insulin resistance associated with regional atrophy in Alzheimer's disease. Hum Brain Mapp 38: 1933-1940. [Crossref]


