Alternative therapy with medical devices using high-voltage electric potential (HELP) to generate an electric field (EF) is common in Japan. In a previous study, HELP exposure-induced upregulation of N-palmitoyl serine (Palmi-Ser) and N-oleoyl serine (Oleo-Ser) in the plasma of healthy individuals was observed. Recently, we found that Oleo-Ser-stimulated fatty acid-binding protein 1 (FABP1) mRNA expression is sensitive to the peroxisome proliferator-activated receptor-alpha (PPAR-α) antagonist. To discover potential pharmacological targets, an in silico docking simulation of Palmi-Ser or Oleo-Ser with ligand-binding domain (LBD) of NR4A2 was investigated. The binding energies of the LBD of NR4A2 were -8.139 and -8.329 kcal/mol for Palmi-Ser and Oleo-Ser, respectively. Palmi-Ser formed hydrogen bonds with Glu-445, Thr-568, and Thr-595. Alternatively, Oleo-Ser formed hydrogen bonds with Thr-567 and Thr-595. In human neuroblastoma SH-SY5Y cells, Palmi-Ser significantly increased NR4A2 mRNA expression. Under these conditions, Oleo-Ser had no effect on NR4A2 mRNA expression. In the Drosophila model of human α-synuclein-expressed Parkinson’s disease, flies treated with Palmi-Ser increased survival rate compared to the vehicle control. Under these conditions, Oleo-Ser had no effect. Palmi-Ser also induced autophagy in human neuroblastoma SH-SY5Y cells. These findings provide new insights into molecular mechanisms of the health benefits for EF therapy through endogenous lipid-derived metabolites.
NR4A2, palmitoyl serine, N-16:0 serine, parkinson model, autophagy
EF: electric field; HELP: high-voltage electric potential; LBD: ligand-binding domain; Oleo-Ser: N-oleoyl serine, N-18:1 serine; Palmi-Ser: N-palmitoyl serine, N-16:0 serine; NR4A2: nuclear receptor subfamily 4 group A member 2; Nurr1: nuclear receptor related-1 protein; PG: prostaglandin; PPAR-α: peroxisome proliferator-activated receptor-alpha
High-voltage electric field (EF) therapy may be effective in treating shoulder stiffness, headache, insomnia, and chronic constipation [1-15]. Although EF therapy was discovered about 90 years ago, the molecular mechanisms associated with its health benefits remain elusive. The Ministry of Health, Labor, and Welfare in Japan approved a therapeutic device that exposes the human body to high-voltage electric potential (HELP) [1-15]. A review of the literature suggests that HELP exposure may be an alternative therapy for several conditions. It was previously reported that HELP exposure-induced upregulation of Palmi-Ser, Oleo-Ser, oleoylethanolamide, 9-hydroxyoctadecadienoic acid (HODE), 13-HODE, lysophosphatidylethanolamine-20:4 (LysoPE-20:4), lysophosphatidylethanolamine-22:6 (LysoPE-22:6), and lysophosphatidylcholine-22:4 (LysoPC-22:4) levels in the plasma of healthy individuals using metabolome analysis [10,12,13,15]. Endogenous lipid-derived metabolites have been suggested as candidate molecules representing the interface between symptoms and electroceutical target proteins [8-15]. Furthermore, defining the molecular interaction between endogenous metabolites and target proteins is essential for understanding the unknown pharmacological effects of human metabolites [16-19]. A recent study by Rajan et al. reported that the crystal structure of lipid-derived metabolites, including prostaglandin PGA2- and PGA1-bound NR4A2-LBD, had been obtained [20,21]. Therefore, it was hypothesized that endogenous lipid-derived metabolites, such as Palmi-Ser and Oleo-Ser may be interacting with NR4A2 as an endogenous ligand. In this study, an in-silico docking simulation was conducted to explore the interactions between Palmi-Ser/Oleo-Ser and NR4A2. The effects of Palmi-Ser and Oleo-Ser on NR4A2 mRNA expression in human neuroblastoma SH-SY5Y cells, and survival rate in the Drosophila model of human α-synuclein-expressed Parkinson’s disease were examined.
Docking study on the NR4A2
The X-ray crystal structure of NR4A2 (also known as Nurr1) complex with PGA2 (5YD6; Protein Data Bank Japan) was used for molecular docking. The docking study of Palmi-Ser/Oleo-Ser binding to the target protein of the hNR4A2 model structure was conducted using the AutoDock Vina docking software (Dr. Oleg Trott, The Scripps Research Institute, CA, USA) [22]. The docking experiment was conducted five times and yielded 100 candidate conformations.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Human neuroblastoma SH-SY5Y cells (ECACC, UK) were cultured in Dulbecco’s modified Eagle’s medium: nutrient mixture F-12 (DMEM/F-12; Thermo Fisher Scientific,Waltham, MA, USA) supplemented with penicillin-streptomycin (Nakarai Tesque, Kyoto, Japan) and 10 % fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA) in a humidified atmosphere of 5% CO2 at 37°C, as described previously [23,24]. SH-SY5Y cells (20,000 cells/well) were subcultured and seeded on 96-well plates (Sumitomo bakelite, Tokyo, Japan). Rat adrenal phaeochromocyte PC-12 cells (ECACC, UK) were cultured in DMEM (low glucose; Nakarai Tesque, Kyoto, Japan), supplemented with penicillin-streptomycin (Nakarai Tesque, Kyoto Japan), 5% FBS, 10% heat-inactivated horse serum (Thermo Fisher Scientific, Waltham, MA, USA) in a humidified atmosphere of 5% CO2 at 37°C. PC-12 cells (10,000 cells/well) were subcultured and seeded on collagen IV-coated 96-well plates (Corning Inc., Corning, NY, USA). After 24 h of incubation, the cells were treated with Oleo-Ser or Palmi-Ser for two hours. The cells treated with vehicle (0.1% ethanol) were used as the control. qRT-PCR was conducted as previously described [8,11,15]. Total RNA isolation and TaqMan-based qRT-PCR was conducted using a FastLane cell probe kit (Qiagen, Valencia, CA, USA) in a Light Cyler 96 system (Roche Applied Science, Mannheim, Germany). Gene expression levels were calculated using the delta-delta-Ct method. Data were presented as relative expression units after normalization to the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene. TaqMan probes were bought from Thermo Fisher Scientific, MA, USA [NR4A2 (Hs01117527_g1), GAPDH (Hs02786624_g1), NR4A1 (Rn00570936_m1), GAPDH (Rn01775763_g1)].
Lifespan assay in the Drosophila model of human α-synuclein-expressed Parkinson’s disease
Fly stocks were maintained at 25°C on standard cornmeal-agar-yeast food (cornmeal 8%, glucose 5%, yeast 5%, agar 0.64%). Transgenic strain UAS-G51D carrying human α-synuclein mutant gene with upstream regulatory sequences (UAS) to derive the causative gene for familial Parkinson’s disease. Neuron-specific driver line elav-gal4 was interbred with UAS-G51D to generate Parkinson’s disease model animal. Each medium was prepared using Formula 4 -24 Blue Drosophila Medium (Carolina Biological Supply Co., Burlington, NC, USA) containing 2-mg/mL dry yeast and 50-µM of each chemical. Lifespan assay was conducted as described previously [25-27]. Male flies of UAS-G51D/elev-gal4 strain were divided into multiple groups of 18-21 individuals each. The flies were housed at a density of 8-7 in a vial containing foods and chemicals and transferred to another vial with fresh medium every three or four days. The survival rate of Drosophila in each group was determined by counting the living flies. The flies were considered dead when they did not move despite agitation [28]. To test for significance, a log-rank test was used by employing Excel Add-in from the addenda of 4Step Excel statics, 4th edition (Yanai hisae, 2015, OMS Publishing Inc.)
Cyto-ID assay
The Cyto-ID Autophagy detection kit (Enzo Life Sciences, Farmingdale, NY, USA) was used according to the manufacturer’s instruction. Briefly, SH-SY5Y cells (10,000 cells/well) were treated using the culture medium containing Palmi-Ser, and were added to the well. After 24h incubation, autophagy was detected using red fluorescent dots derived from Cyto-ID Dye existing in the cytoplasmic region. Imaging analysis was conducted using Operetta CLS (Perkin Elmer, Waltham, MA, USA).
Chemicals
Palmi-Ser and rapamycin were bought from Sigma-Aldrich (St. Louis, MO, USA). Oleo-Ser and PGA2 were bought from Cayman Chemical (Ann Arbor, MI, USA).
Statistical analysis
Data were analyzed using Welch’s t-test. A probability (p) value < 0.05 was considered statistically significant.
Docking simulation of Palmi-Ser and Oleo-Ser with LBD of NR4A2
To determine whether Palmi-Ser and Oleo-Ser were involved with LBD of NR4A2, an in silico docking simulation of NR4A2 was examined next (Figure 1). Since the protein database (PDB) has four 3D structures of hNR4A2 (1OVL, 5YD6, 5Y41, and 6DDA), an overlay of these four PDB structures was first observed (Figure 2). Small molecule ligands of the co-crystal structures 5YD6 (NR4A2 LBD bound to PGA2), 5Y41 (NR4A2 LBD bound to PGA1), and 6DDA (NR4A2 LBD bound to 5, 6-dihydroxyindole) were found to interact with helix-12. Since 1OVL is not a crystal structure containing ligands, docking simulations of 5YD6, 5Y41, and 6DDA were investigated. Using chloroquine, which had already undergone docking analysis as an agonist for NR4A2 [29], the best (or mean) scores were -8.496 (-8.087), -8.325 (-7.764), and -6.775 (-6.562) kcal/mol for 5YD6, 5Y41, and 6DDA, respectively. Subsequent analysis was conducted using the PDB structure of 5YD6, which had a good docking score for chloroquine. An in silico docking of Oleo-Ser and Palmi-Ser in the LBD of NR4A2 was examined using AutoDock Vina software. The number of output poses were set to 20, with 100 candidate conformations. Palmi-Ser exhibited good binding energy of -8.139 kcal/mol (Table 1). Palmi-Ser formed hydrogen bonds with Glu-445, Thr-567, and Thr-595 (Figure 2) (Table 1). Alternatively, Oleo-Ser exhibited good binding energy of -8.329 kcal/mol (Table 1). Oleo-Ser formed hydrogen bonds with Thr-567 and Thr-595 (Figure 2) (Table 1).
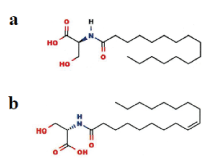
Figure 1. Chemical structure of Palmi-Ser and Oleo-Ser. (a) Palmi-Ser (b) Oleo-Ser
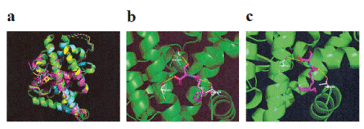
Figure 2. In silico molecular docking of Palmi-Ser and Oleo-Ser with LBD of NR4A2. (a) Superposition of the NR4A2 LBD (PDB code: 1OVL) in green, with NR4A2 LBD (PDB code: 5YD6) in cyan, NR4A2 LBD (PDB code: 5Y41) in magenta, and NR4A2 LBD (PDB code: 6DDA) in yellow. (b) Binding mode of Palmi-Ser in modeling of NR4A2 (PDB code: 5YD6). Magenta represents Palmi-Ser and white represents the amino acid making up the pocket. The yellow dashed line indicates hydrogen bonding. The hydroxyl group of Palmi-Ser forms a hydrogen bond with the Glu-445 oxygen atom. The oxygen atom of Palmi-Ser forms a hydrogen bond with the Thr-567 hydroxyl group. The oxygen atom of Palmi-Ser forms a hydrogen bond with the Thr-595 hydroxyl group. (c) Binding mode of Oleo-Ser in modeling of NR4A2 (PDB code: 5YD6). Magenta represents Oleo-Ser and white represents the amino acid making up the pocket. The yellow dashed line indicates hydrogen bonding. The oxygen atom of Oleo-Ser forms a hydrogen bond with the Thr-567 hydroxyl group. The oxygen atom of Oleo-Ser forms a hydrogen bond with the Thr-595 hydroxyl group
Table 1. Docking score and key interacting residues of hNR4A2 (5YD6)
Ligand |
Docking score (kcal/mol) |
Interactive residues |
|
|
| |
|
Palmi-Ser |
-8.139 |
Glu-445, Thr-567, and Thr-595 |
|
|
Oleo-Ser |
-8.329 |
Thr-567, and Thr-595 |
|
|
Effects of Palmi-Ser and Oleo-Ser on NR4A2 mRNA expression in human neuroblastoma SH-SY5Y cells
Chloroquine, an agonist for NR4A2, is known to regulate NR4A2 mRNA through direct binding to NR4A2-LBD [29]. In addition, PG analogues such as PGA1, PGE1, and PGE2 are known to activate transcriptional function [21,29,30]. Therefore, we examined the effects of Palmi-Ser and Oleo-Ser on NR4A2 mRNA expression using qRT-PCR were examined (Figure 3). In human neuroblastoma SH-SY5Y cells, Palmi-Ser increased NR4A2 mRNA expression in SH-SY5Y cells (Figure 3) (Palmi-Ser 1-µM:1.27-fold, p=0.0408; Palmi-Ser 10-µM:1.24-fold, p=0.0486). However, under these conditions, treatments of Oleo-Ser did not affect NR4A2 mRNA expression (Figure 3) (Oleo-Ser 1-µM:0.84-fold, p=0.3991; Oleo-Ser 10-µM:1.12-fold, p=0.6521).
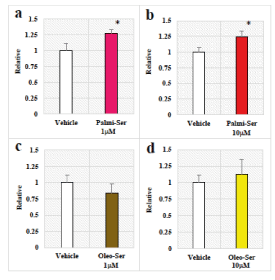
Figure 3. Effects of Palmi-Ser and Ole-Ser on NR4A2 mRNA expression in human neuroblastoma SH-SY5Y cells. (a) Effect of Palmi-Ser (1-mM) on NR4A2 mRNA expression. Data are expressed as mean ± SE (n=18). (b) Effect of Palmi-Ser (10-mM) on NR4A2 mRNA expression. Data are expressed as mean ± SE (n=31). (c) Effect of Oleo-Ser (1-mM) on NR4A2 mRNA expression. Data are expressed as mean ± SE (n=8). (d) Effect of Oleo-Ser (10-mM) on NR4A2 mRNA expression. Data are expressed as mean ± SE (n=8). * p < 0.05 compared with the vehicle
To determine the specificity of Palmi-Ser, next the effect of Palmi-Ser on NR4A1 (also known as NGFI-B) mRNA expression in rat adrenal phaeochromocyte PC-12 cells was examined. Exposure of the cells to 10-µM Palmi-Ser did not affect NR4A1 mRNA expression (0.98-fold, p=0.859).
The effect of Palmi-Ser on survival rate in the Drosophila model of human α-synuclein-expressed Parkinson’s disease
The effects of Palmi-Ser and Oleo-Ser on survival rate in the transgenic Drosophila model of human α-synuclein-expressed Parkinson’s disease were further examined (Figure 4). Treatment with Palmi-Ser had significantly increased survival rate compared to vehicle control in the transgenic Drosophila model of human α-synuclein-expressed Parkinson’s disease (Figure 4) (p=0.0004, log-rank test). In contrast, the treatment of Oleo-Ser did not affect so much for survival rate compared to vehicle control in the transgenic Drosophila model of human α-synuclein-expressed Parkinson’s disease (Figure 4) (p=0.12, log-rank test). Under the same conditions, PGA2, one of agonist candidates for NR4A2, had significantly increased survival rate compared to vehicle control as expected in the transgenic Drosophila model of human α-synuclein-expressed Parkinson’s disease (Figure 4) (p=0.04, log-rank test).
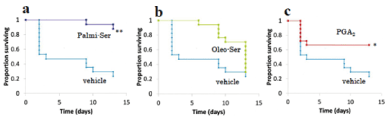
Figure 4. The effect of Palmi-Ser on survival rate in the Drosophila model of human a-synuclein-expressed Parkinson’s disease. (a) The effect of Palmi-Ser (50-mM) on survival rate in the Drosophila model of human a-synuclein-expressed Parkinson’s disease (n=18). (b) Effect of Oleo-Ser (50-µM) on survival rate in the Drosophila model of human a-synuclein-expressed Parkinson’s disease (n=18). (c) Effect of PGA2 (50-mM) on survival rate in the Drosophila model of human a-synuclein-expressed Parkinson’s disease (n=18). * p < 0.05 compared with the vehicle. ** p < 0.01 compared with the vehicle
The effect of Palmi-Ser on autophagy in human neuroblastoma SH-SY5Y cells
Because autophagy is an integral mediator of lifespan extension [31], we hypothesized that increased survival rate may be linked to changes in autophagy. Therefore, the effect of Palmi-Ser on autophagy was examined next using Cyto-ID-based fluorescence spectrophotometric assay (Figure 5). Palmi-Ser dose-dependently increased Cyto-ID-stained autophagic compartments (Palmi-Ser 5-µM: 1.12-fold, p=0.228; Palmi-Ser 15-µM: 1.20-fold, p=0.045; Palmi-Ser 50-µM: 1.18-fold, p=0.071). Under these conditions, a well-known autophagy activator rapamycin (20-µM) increased Cyto-ID-stained autophagic compartments (rapamycin 20-µM: 2.3-fold, p=2.5E-7).
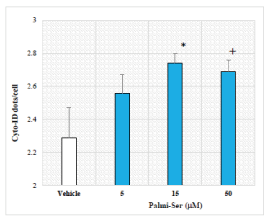
Figure 5. The effect of Palmi-Ser on autophagy in human neuroblastoma SH-SY5Y cells. Effect of Palmi-Ser on Cyto-ID dots/cell in human neuroblastoma SH-SY5Y cells. Data are expressed as mean ± SE (n=5). + p < 0.1 compared with the vehicle. * p < 0.05 compared with the vehicle
Since the first report on the crystal structure of apoNR4A2 (1OVL), the NR4A2 function is believed to be independent of the binding ligand [32]. Thus far, there is little evidence that these ligands directly activate the NR4A2. Most published studies were conducted using the orphan nuclear receptor NR4A1 (Nur77) or neuron-derived orphan receptor (NOR1; NR4A3) LBD. The amino acid sequences of the LBD between NR4A1 with NR4A2 and NR4A3 show a sequence similarity of 58%-65% [33]. Lakshmi et al. reported activation of the NR4A1 by PGA2 [34], while Kagaya et al. reported that PGA2-stimulated NR4A3 activation by binding directly to the NR4A3’s LBD [35]. Alternatively, considerable evidence for NR4A2 activation by the anti-malarial drugs such as amodiaquine and chloroquine suggested the presence of a ligand-binding pocket within the LBD of NR4A2 [29,36]. Furthermore, the most recent reports on a dopamine metabolite DHI and PG family (PGE1, PGA1, and PGA2) for NR4A2’s LBD have triggered and shed light on endogenous ligands [20,21,37]. In this study, the docking simulation with NR4A2 showed that Palmi-Ser has a good binding affinity value (-8.139 kcal/mol). Palmi-Ser binding to NR4A2 was stabilized through the formation of hydrogen bonds with Glu-445, Thr-567, and Thr-595. On the other hand, the docking simulation with NR4A2 showed that Oleo-Ser has a good binding affinity value (-8.329 kcal/mol). Oleo-Ser binding to NR4A2 was stabilized though the formation of hydrogen bonds with Thr-567 and Thr-595. In key interacting residues, a previous study using two-dimensional 1H-15N Heteronuclear Single Quantum Correlation spectra of the 15N-labeled NR4A2-LBD with chloroquine-binding pockets reported hydrogen bonding to Thr-595 in C-terminal helix-12 [29]. Another study on PGA1/PGA2-binding pockets of NR4A2 reported hydrogen bonding to Glu-445, Thr-567, and Thr-595 [20,21]. Additionally, Bruning et al. reported that a hydrogen bond between the carboxyl group of Glu-445 and the NH of DHI in docking simulation of 6DDA [37]. These findings indicate that Palmi-Ser may act as endogenous ligands for NR4A2’s LBD.
Park et al. reported that an increase in NR4A2 mRNA induced by chloroquine, an agonist for NR4A2 [29]. The results of present study showed that Palmi-Ser (1µM) induced an approximately 1.3-fold increase in NR4A2 mRNA expression in a human neuroblastoma SH-SY5Y cell line. The data indicate that Palmi-Ser may act as an endogenous agonist for NR4A2’s LBD. In contrast, Oleo-Ser had no effect on NR4A2 mRNA expression. Thus Oleo-Ser may act as a weak partial agonist for NR4A2. Unfortunately, there is no commercially available antagonist of NR4A2, the effect of NR4A2 antagonist was not performed in this study. However, Palmi-Ser reported to affect PPARα, transient receptor potential vanilloid 1, and cannabinoid receptor [15,38], raising the possibility that these receptors may be targets for Palmi-Ser.
NR4A2 has emerged as a promising therapeutic target for Parkinson’s disease [39,40]. This study showed that Palmi-Ser reduces mortality in the Drosophila model of α-synuclein-induced Parkinson’s disease. The data suggests that Palmi-Ser may act as an agonist for the Drosophila NR4A nuclear receptor, DHR38. In this study, it was demonstrated that Palmi-Ser induced autophagy in human neuroblastoma SH-SY5Y cells. Interestingly, Winslow et al. reported that the inhibition of autophagy by α-synuclein overexpression might explain the pathology of Parkinson’s disease [41], while Sato et al. reported that the loss of normal autophagy in dopaminergic neurons causes Lewy body pathology in Parkinson’s disease [42]. In addition, Kaji et al. reported that α-synuclein accumulation–induced changes in autophagic indicators such as p62 and LC3-2/LC3-1 in oligodendrocyte precursor cells [43], while Uemura et al. reported that α-synuclein accumulation in axonal swellings containing autophagosomes [44]. An acute EF exposure (9 kV/electrode + 9 kV/electrode, 30 min)-induced increase in the levels of Palmi-Ser (approximately 1.42-fold) in plasma from healthy humans was previously shown [15]. Therefore, future studies may be interesting to evaluate the possible effects of Palmi-Ser or EF therapy on dementia with Levy bodies in Parkinson’s disease [45-47].
Molecular docking of Palmi-Ser was observed in the model of NR4A2 LBD in silico. Palmi-Ser significantly increased survival rate in the Drosophila model of human α-synuclein-expressed Parkinson’s disease in vivo. Palmi-Ser induced autophagy in human neuroblastoma SH-SY5Y cells in vitro. These findings provide insights into the molecular mechanisms behind the health benefits induced by the HELP device.
YN-Y, TN, AH, and HH are employees of Hakuju Institute for Health Science Co., Ltd. TY is employed by Acel Inc. EK is employed by Intage Healthcare Inc, and HK and NI are employed by Foundation for Advancement of International Sciences. All other authors have no competing interests.
YN-Y and TN contributed equally to this work. YN-Y designed and supervised the research project, and wrote the manuscript. YN-Y, TN, TY, and HK performed biological experiments. EK performed molecular modeling. AH, HH, and NI helped supervise the project. All authors have read and approved the final version of the manuscript.
We thank Dr. Makoto Kikuchi (Professor Emeritus, National Defense Medical College, Japan) for his encouragement.
- Hara H (1961) On the effect of AC. electrostatic high voltage potential load upon the blood-electrolytes (in Japanese). Niigata Medical J 75: 265-273.
- Ito F, Furuya K (1981) The effect of high voltage alternating current upon a human body the change of blood pressure, endocrine system and serum lipids (in Japanese). J Jpn Sci Balneol Climatol Phys Med 45: 6-17.
- Isaka K, Nishimura R, Arase S, Takiwaki H, Osaki K, et al. (1998) Dosimetry and exposure experiments for extremely low frequency high-tension electric field therapy. EMC ’98 Rome International Symposium on Electromagnetic Comoatibility D: 204-207.
- Nawarat S, Iomsai K, Jantanam P, Kauengtip Y (1999) Effects of electrical Healthtron on curing of non-communicable diseases: Case study of Banlad hospital Phetchaburi province (in Thai). Region 4 Medical J 18: 139-149.
- Siripanichgon K, Otrakul A, Suparp J, Sirikulchayanonta C, Charupoonphol P (2000) Clinical observation of Healthtron therapy (in Thai). J Public Health (Bangkok) 30: 19-29.
- Sirikulchayanonta C, Siripanichgon K, Otrakul A, Suparp J, Charupoonphol P (2001) The effect of Healthtron on serum lipid levels among the middle-aged: Preliminary report. J Public Health (Bangkok) 31: 63-70.
- Ito F, Ohsaki K, Takahasi K, Hara H (2005) The effects of electric field therapeutic device (Healthtron) on the stiffness in the neck and shoulder area – changes in subjective symptoms, blood circulation and the autonomic nervous system (in Japanese). J Jpn Sci Balneol Climatol Phys Med 68: 110-121.
- Nakagawa-Yagi Y, Hara H, Fujimori T, Yamaguchi T, Midorikawa A, et al. (2014) Non-targeted human plasma metabolomics reveals the changes in oleoylethanolamide, a lipid-derived signaling molecule, by acute exposure of electric field. Integr Mol Med 1: 29-37
- Nakagawa-Yagi Y, Hara H, Yoshida Y, Midorikawa A, Hara A (2015) Discovery of a novel effect of electric field exposure on human plasma beta-endorphin and interleukin-12 levels: Insight into mechanisms of pain alleviation and defense against infection by electric field therapy. Integr Mol Med 2: 200-204.
- Nakagawa-Yagi Y, Hara H, Nakagawa F, Sato M, Hara A (2016) Acute exposure to an electric field induces changes in human plasma 9-HODE, 13-HODE, and immunoreactive substance P levels: Insight into the molecular mechanisms of electric field therapy. Integr Mol Med 3: 600-605
- Nakagawa-Yagi Y, Hara H, Tsuboi H, Abe J, Hara A (2016) Effect of 3-hydroxybutyrate, an endogenous histone deacetylase inhibitor, on FOXO3A mRNA expression in human epithelial colorectal Caco-2 cells: Insight into the epigenetic mechanisms of electric field therapy. Integr Mol Med 3: 764-768.
- Nakagawa-Yagi Y, Hara H, Nakanishi H, Tasaka T, Hara A (2017) Acute exposure to an electric field induces changes in human plasma lysophosphatidylcholine (lysoPC)-22:4 levels: Molecular insight into the docking of lysoPC-22:4 interaction with TRPV2. Integr Mol Med 4: 1-7.
- Nakagawa-Yagi Y, Hara H, Nakanishi H, Kanai C, Hara A (2017) Molecular insight into the docking of lysophosphatidylethanolamine (lysoPE)-22:6 interaction with GPR119: Acute exposure to an electric field induces changes in human plasma lysoPE-22:6 and lysoPE-20:4 levels. Integr Mol Med 4: 1-7.
- Nakagawa-Yagi Y, Hara H, Kanai C, Sato M, Hara A (2018) Acute electric field downregulates human plasma immunoreactive interleukin-6 and -1 β levels: Molecular mechanisms underlying inflammation alleviation through electric field therapy. Integr Mol Med 5: 1-6.
- Nakagawa-Yagi Y, Hara H, Kanai C, Sato M, Hara A (2019) Targeted lipidomics reveals changes in N-acyl serines by acute exposure to an electric field: Molecular insights into the docking of N-18:1 serine interaction with TRPV1 or PPAR- α. Integr Mol Med 6: 1-8.
- Nakagawa-Yagi Y, Ogane N, Inoki Y, Kitoh N (1996) The endogenous estrogen metabolite 2-methoxyestradiol induces apoptotic neuronal cell death in vitro. Life Sci 58: 1461-1467. [Crossref]
- Tonack S, Tang C, Offermanns S (2013) Endogenous metabolites as ligands for G protein-coupled receptors modulating risk factors for metabolic and cardiovascular disease. Am J Physiol Heart Circ Physiol 304: H501-H513.
- Newman JC, Verdin E (2014) Ketone bodies as signaling metabolites. Trends Endocrinol Metab 25: 42-52.
- Piomelli D, Sasso O (2014) Peripheral gating of pain signals by endogenous lipid mediators. Nat Neurosci 17: 164-174.
- Rajan S, Toh HT, Lim KH, Yoon HS (2019) Structure of Nurr1 bound to cyclopentenone prostaglandin A2 and its mechanism of action in ameliorating dopaminergic neurodegeneration in Drosophila. PDB ID 5YD6.
- Rajan S, Jang Y, Kim CH, Kim W, Toh HT, et al. (2020) PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function. Nat Chem Biol 16: 876-886.
- Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31: 455-461.
- Nakagawa-Yagi Y, Saito Y, Takada Y, Takayama M (1991) Carbachol enhances forskolin-stimulated cyclic AMP accumulation via activation of calmodulin system in human neuroblastoma SH-SY5Y cells. Biochem Biophys Res Commun 178: 116-123.
- Nakagawa-Yagi Y (1994) Induction of apoptotic cell death in differentiating in human neuroblastoma SH-SY5Y cells by colchicine. Biochem Biophys Res Commun 199: 807-817.
- Feany MB, Bender WW (2000) A Drosophila model of Parkinson’s disease. Nature 404: 394-398.
- Ito K, Kawasaki H, Suzuki T, Takahara T, Ishida N (2017) Effects of Kamikihito and Unkei-to on sleep behavior of wild type and Parkinson model in Drosophila. Front Psychiatry 8: 132.
- Mohite GM, Dwivedi S, Das S, Kumar R, Paluri S, et al. (2018) Parkinson's disease associated α-synuclein familial mutants promote dopaminergic neuronal death in Drosophila melanogaster. ACS Chem Neurosci 9: 2628-2638. [Crossref]
- Kawasaki H, Suzuki T, Ito K, Takahara T, Goto-Inoue N, et al. (2018) Minos-insetion mutant of the Drosophila GBA gene homologue showed abnormal phenotypes of climbing ability, sleep and life span with accumulation of hydroxy-glucocerebroside. Gene 614: 49-55.
- Park TY, Jang Y, Kim W, Shin J, Toh HT, et al. (2019) Chloroquine modulates inflammatory autoimmune responses through Nurr1 in autoimmune diseases. Sci Rep 9: 15559.
- Holla VR, Mann JR, Shi Q, DuBois RN (2006) Prostaglandin E2 regulates the nuclear receptor NR4A2 in colorectal cancer. J Biol Chem 281: 2676-2682.
- Madeo F, Zimmermann A, Maiuri MC, Kroemer G (2015) Essential role for autophagy in life span extension. J Clin Invest 125: 85-93. [Crossref]
- Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, et al. (2003) Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 423: 555-560.
- Kurakula K, Koenis DS, van Tiel CM, de Vries CJM (2014) NR4A nuclear receptors are orphans but not lonesome. Biochim Biophys Acta 1843: 2543-2555.
- Lakshmi S, Reddy AT, Banno A, Reddy RC (2019) Molecular, chemical, and structural characterization of prostaglandin A2 as a novel agonist for Nurr77. Biochem J 476: 2757-2767.
- Kagaya S, Ohkura N, Tsukada T, Miyagawa M, Sugita Y, et al. (2005) Prostaglandin A2 acts as a transactivator for NOR1 (NR4A3) within the nuclear receptor superfamily. Biol Pharm Bull 28: 1603-1607.
- Kim CH, Han BS, Moon J, Kim DJ, Shin J, et al. (2015) Nuclear receptor Nurr1 agonists enhance its dual functions and improve behavioral deficits in an animal model of Parkinson’s disease. Proc Natl Acad Sci USA 112: 8756-8761.
- Bruning JM, Wang Y, Oltrabella F, Tian B, Kholodar SA, et al. (2019) Covalent modification and regulation of the nuclear receptor Nurr1 by a dopamine metabolite. Cell Chem Biol 26: 674-685. [Crossref]
- Mann A, Cohen-Yeshurun A, Trembovler V, Mechoulam R, Shohami E (2016) Are the endocannabinoid-like compopunds N-acyl aminoacids neuroprotective after traumatic brain injury? J Basic Clin Physiol Pharmacol 27: 209-216.
- Chu Y, Le W, Kompoliti K, Jankovic J, Mufson EJ, et al. (2006) Nurr1 in Parkinson’s disease and related disorders. J Comp Neurol 494: 495-514.
- Decressac M, Volakakis N, Björklund A, Perlmann T (2013) Nurr1 in Parkinson disease - from pathogenesis to therapeutic potential. Nat Rev Neurol 9: 629-636.
- Winslow AR, Chen CH, Corrochano S, Acevedo-Arozena A, Gordon DE, et al. (2010) α-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol 190: 1023-1037.
- Sato S, Uchihara T, Fukuda T, Noda S, Kondo H, et al. (2018) Loss of autophagy in dopaminergic neurons causes Lewy pathology and motor dysfunction in aged mice. Sci Rep 8: 2813.
- Kaji S, Maki T, Kinoshita H, Uemura N, Ayaki T, et al. (2018) Pathological endogenous a-synuclein accumulation in oligodendrocyte precursor cells potentially induces inclusions in multiple system atrophy. Stem Cell Reports 10: 356-365.
- Uemura N, Koike M, Ansai S, Kinoshita M, Ishikawa-Fujiwara T, et al. (2015) Viable neuronopathic Gaucher disease model in Medaka (Oryzias latipes) displays axonal accumulation of alpha-synuclein. PLoS Genet 11: e1005065.
- Kosaka K, Mehraein P (1979) Dementia-Parkinsonism syndrome with numerous Lewy bodies and senile plaques in cerebral cortex. Arch Psychiatr Nervenkr 226: 241-250.
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, et al. (1997) α-Synuclein in Lewy bodies. Nature 388: 839-840.
- Kosaka K (2014) Lewy bodies disease and dementia with Lewy bodies. Proc Jpn Acad Ser B Phys Biol Sci 90: 301-306.
Editorial Information
Editor-in-Chief
Prof. Hamid Yahya Husain
Dubai Health Authority, Dubai
Article Type
Research Article
Publication history
Received date: October 18, 2021
Accepted date: November 1, 2021
Published date: November 10, 2021
Copyright
©2021 Yuzo Nakagawa-Yagi, Takaki Nedachi, Haruhisa Kawasaki, Takahiro Yamaguchi, Enzo Kawasaki, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation
Yuzo Nakagawa-Yagi (2021) Molecular insights into the docking of N-palmitoyl serine (Palmi-Ser) interaction with the nuclear receptor subfamily 4 group A member 2 (NR4A2): Effect of Palmi-Ser on survival rate in the Drosophila model of Parkinson's disease. Med Case Rep Rev, 2021 DOI: 10.15761/MCRR.1000173





