Abstract
Multi Omics by LC-MS/MS is useful tool to search the ligands to control transcription of pharmaceutical active proteins such TNF, IL-1 and peroxisome.
Key words
nuclear receptor, multi omics, mass spectrometry, drug discovery
How to search the small molecule ligands of the nuclear receptors to control transcription of pharmaceutical active properties using multi omics by LC-MS/MS
Osaka university shimadzu analytical innovation research laboratory is developing multi omics (MO) package on Garuda platform [1] that have statist analysis (principal components analysis (PCA)) and correlation analysis (the gene ontology analysis and pathway analysis) to treat LC-MS/MS data expressed in KEGG map in Figure 1. We believe that MO that is useful tool to discover a new drug as the interacted inhibitors of the small molecule ligands of nuclear receptor (NR) bound to promoter DNA to control transcription of pharmaceutical active proteins. NR has both L- domain binding to the small molecule ligands and D-domain binding to the promoter DNA as transcription factor. The human 48 NR contain 40 orphan receptors and 8 endocrine receptors that bind steroid hormones ligands produced from cholesterol by metabolism with CYP11, 17, 19 and 21 etc. Some ligands of orphan NR are unknown yet in Figure 2.
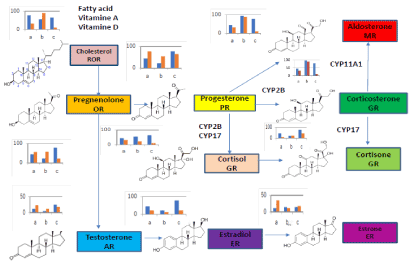
Figure1. PCA Result of LC-MS/MS data expressed in KEGG map of steroid metabolism
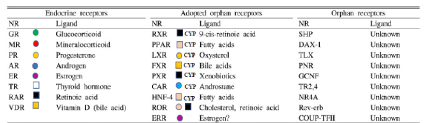
Figure 2. Ligands and nuclear receptor as transcription factor
We have developed FK506 NF-AT transcription inhibitor [2], FK228 PPAR inhibitor [3], NF-kB Decoy oligo nucleotide (ON) [4,5] and AO85 Exon Skipping ON [6,7] that control transcription. FK506 and NF-κB decoy ON inhibits expression of inflammatory cytokines, such as TNF and IL-1 which trigger inflammation cascades to dermatitis. Antisense ON 18-mer 20-OMeRNA/ENA chimera (AO85) had the most potent activity for inducing exon 45 skipping of the dystrophin reading frame to allow production of an internally deleted dystrophin protein with functional benefit for DMD patients who have out-of-frame deletions. We presented plasma, urine, tissue MO and identified many transcription factors bound to a new promoter DNA of the platelet factor 4 [8] or a known promoter DNA of CYP3A4 by nanospray LC-MS/MS after in-gel tryptic digestion of spots SDS page electrophoresis. We will describe these unpublished studies as Articles in this journal continuously.
Acknowledgements
We would express our special thanks to Managing Editor Sandy Williams for encouragement and introduction to Trends in Research, which aims to open and share various new research idea and data for human benefits and public good.
Conflicts of interest
The authors declare no conflict of interest.
References
- Ghosh S, Matsuoka Y, Asai Y, Hsin KY, Kitano H (2011) Software for systems biology: From tools to integrated platforms. Nature Rev Genet 12: 821-832.
- Tozuka Z, Kaneko H, Shiraga T, Mitani Y, Beppu M, et al. (2003) Strategy for structural elucidation of drugs and drug metabolites using (MS)n fragmentation in an electrospray ion trap. J Mass Spectrom 38: 793-808. [Crossref]
- Tozuka Z, Strupat WM, Shiraga T, Ishimura R, Hashimoto T (2005) LTQ FT MS accurate mass and SRM data dependent exclusion MSn measurements for structure determination of FK228 and its metabolites. J Mass Spectrum Soc Jpn 53: 89-99.
- Wada K, Morishita R (2010) Clinical development of NF?B decoy oligodeoxy nucleotides. J-STAGE, Drug Delivery System 25: 579-589.
- Shioyama S, Tozuka Z. Analysis of metabolites in plasma afteriv dosing of NF?B decoy oligodeoxy nucleotides. JCL Bioassay Corporation Internal report JCL059132
- Dwianingsih EK, Malueka RG, Nishida A, Itoh K, Lee T, et al. (2014) A novel splicing silencer generated by DMD exon 45 deletion junction could explain upstream exon 44 skipping that modifies dystrophinopathy. J Hum Genet 59: 423-429. [Crossref]
- Tozuka Z. Validation study, Quantitative analysis of AO85 by ESI LC/MS. Sekisui Medical Co., Ltd. Internal report AL-4450.
- Okada Y, Matsuura E, Tozuka Z, Nagai R, Watanabe A, et al. (2004) Upstream stimulatory factors stimulate transcription through E-box motifs in the PF4 gene in megakaryocytes. Blood 104: 2027. [Crossref]
Editorial Information
Editor-in-Chief
Hiroyuki Aizawa
Aizawa Science Museum
Article Type
Short Communication Article
Publication history
Received: July 02, 2018
Accepted: July 19, 2018
Published: July 23, 2018
Copyright
©2018 Tozuka Z. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation
Tozuka Z, Kunisawa A (2018) Multi Omics by LC-MS/MS to search small molecule ligands of nuclear receptors to control transcription of pharmaceutical active proteins for drug discovery. Trends in Res 1: DOI: 10.15761/TR.1000111
Corresponding author
Zenzaburo Tozuka
Osaka University Shimadzu Analytical Innovation Research Laboratory, 2-1, Yamada-Oka, Suita, Osaka 565-0871, Japan


