Abstract
DNA is highly vulnerable to environmental agents like ultraviolet (UV) light. DNA polymerase η (polη) catalyzes accurate bypass of cyclobutane pyrimidine dimers (CPDs) and suppresses UV-induced mutagenesis. Polη harbors three PCNA-interacting peptides (PIP) boxes, whereas PIP boxes in polη exhibit non-canonical sequences. Here, we established the mutant cells stably expressing polη having consensus sequences of PIP1, PIP3 or PIP2 into polη-deficient cells. We demonstrated that either PIP1, PIP3, or PIP2 consensus mutant polη complements UV sensitivities of XP2SASV3 cells as wild type polη. However, expression of the double PIP consensus mutant of polη results in diminished cell viability. Furthermore, according to mass spectrometry analysis, peptides containing PIP1 and PIP3 of polη are phosphorylated in human cells. We also revealed that phospho-mimetic mutant of polη in PIP1 or PIP3 reduced the complementation activity of wild type, mutations in PIP1/PIP3 of polη further reduced the complementation ability. Based on these results, we proposed a mechanistic model the mutation of polη in its PIP boxes played a dual role in regulating its functions and also in polymerase switching, where consensus mutant polη competes with wild type polη to execute lesion bypass. In contrast, phosphorylation of polη in its PIP1 and PIP3 negatively regulate polη’s function, disturbing the polymerase switching and resulting in replication stress.
Key words
Polη, UV sensitivity, DNA damage, DNA translesion synthesis (TLS)
Genomic DNA in living organisms is continually threatened by DNA damaging agents such as ultraviolet (UV) light. Multiple DNA damage response (DDR) pathways have evolved to maintain the integrity of genomic DNA. Accurate and integral cellular DNA replication is modulated by multiple replication-associated proteins, which is fundamental to preserve genome stability. Furthermore, replication proteins cooperate with multiple DNA damage repair factors to deal with replication stress through mechanisms beyond their role in replication. However, replicative DNA polymerases could not encounter damaged DNA, thus blocking DNA replication fork. If this problem weren’t be resolved, replication fork would be collapsed, resulting in cell death [1,2]. Two major types of DNA lesions produced by UV irradiation are cyclobutane pyrimidine dimers (CPDs) and (6-4) photoproducts (6-4 PPs). 6-4 PPs are repaired rapidly by nucleotide excision repair (NER), whereas CPDs are repaired much more slowly and give rise to stalled DNA replication fork [3,4].
Cells have mechanisms to resolve replication fork stalling, so-called DNA damage tolerance (DDT) pathways such as translesion DNA synthesis (TLS) [5,6]. TLS is carried out by specialized DNA polymerases, called TLS polymerase. One of the TLS polymerases is human DNA polymerase η (polη) [7]. Polη catalyzes efficient and accurate TLS past thymine-thymine CPD (T-T CPD) [8,9]. Disabled polη leads to a hereditary disease xeroderma pigmentosum variant (XPV) which is characterized by a high frequency of skin cancer. However, polη lacks 3’-5’ proofreading exonuclease activities and replicates undamaged DNA with low fidelity. Therefore, polη should be strictly regulated to replicate damaged DNA [10,11].
Proliferating cell nuclear antigen (PCNA) was initially characterized as a DNA sliding clamp for replicative DNA polymerases and an essential component of the eukaryotic chromosomal DNA replisome [12,13]. Subsequent studies have revealed its remarkable ability to interact with multiple partners, which are involved in several metabolic pathways, including cell cycle regulation, TLS, and DNA damage repair [14,15]. It has been revealed that PCNA plays a vital role in regulating polη function [16,17]. PCNA is mono-ubiquitinated at K164 by RAD6-RAD18 E2-E3 complex in response to replication fork stalling [18,19]. Mono-ubiquitinated PCNA increases interacting affinity with polη. Mono-ubiquitination of PCNA plays critical roles in enabling TLS by interacting with polη [20,21]. Numerous PCNA-interacting proteins cooperate with PCNA via their PCNA interacting protein (PIP) box. A typical consensus amino acid sequence of PIP motif is (Q-x-x-(I/L/M)-x-x-(F/Y)-(F/Y)) [22,23]. Polη harbors three PIP boxes, PIP1, PIP3, and PIP2, which exhibit non-canonical sequence and perform weak interaction ability with PCNA [24,25].
Previous research demonstrated that three PIP boxes in polη contribute to two distinct functions, stimulation of DNA synthesis and promotion of PCNA ubiquitination. A deletion mutant carrying the 1-511 region of polη (polηΔC) lacking PIP2 could complement the UV sensitivity of XP2SASV3 cells, which suggested that PIP1, PIP3, and PIP2 exhibit additive and redundant effect in protecting cells from UV irradiation [26].
Moreover, some studies suggested that post-translational modifications (PTMs) of polη involves in TLS regulation, especially phosphorylation modification [27]. Chen et al. demonstrated that inhibiting phosphorylation of S587 and T617 of polη in XP2SASV3 cells reduced UV complementation activity compared with WT, suggesting that phosphorylation of polη could regulate TLS [28]. In addition, phosphorylation of S687 in polη could diminish interaction ability with PCNA, further interrupting normal TLS process. Phospho-deficient mutant S687A reduced the UV complementation ability of polη, indicating that phosphorylation of polη in S687 modulates cellular resistance toward UV irradiation [29]. Moreover, S601 residue of polη is phosphorylated after UV irradiation. S601A mutant polη displayed lower survival after UV irradiation, which enunciating S601 phosphorylation of polη regulating UV damage bypassing ability [11,30]. In addition, some reports also demonstrated that phosphorylation of S601, S687, and S510, S512, and S514 residues are vital for damage bypassing and cell survival after UV irradiation [11]. Peddu and co-workers also demonstrated that phosphorylation of polη in S587, T617, and S601 potentiates its affinity with Ub-PCNA [31]. However, little is known about how phosphorylation of PIP boxes in regulating polη function.
Here we report that PIP boxes in polη are crucial components and exist in different mechanisms via their distinct modifications. We identify novel PIP boxes consensus mutant of polη and phosphorylation sites, which affect polη stability and modulate the TLS process.
Materials and methods
Construction of mutants polη-expressing plasmids
pIRESneo2/FLAG-polη and pIRESneo2/FLAG- polηΔC were used to establish mutant polη expressing plasmid. pIRESneo2/FLAG-polη PIP3 (T477D), pIRESneo2/FLAG- polηΔC PIP1(SSS435-7AAA), pIRESneo2/FLAG- polηΔC PIP1(SSS435-7DDD), pIRESneo2/FLAG- polηΔC PIP3(TTS477-9AAA), pIRESneo2/FLAG- polηΔC PIP3(TTS477-9DDD), pIRESneo2/FLAG- polηΔC PIP1/3(SSS435-7AAA/TTS477-9AAA) and pIRESneo2/FLAG- polηΔC PIP1/3(SSS435-7DDD/TTS477-9DDD) were made by site-directed mutagenesis, using the following primers and verified by sequencing:
PIP3 F: 5’-ACAGCCACTAAGAAAGCACAGACGTCTCTGGAATCATTC-3’
SSS435-7AAA-S: 5’-TTCTGCCTCTGCCCCTGCAGCTGCTACAGACATCACCAGCT-3’
SSS435-7DDD-S: 5’-CTGCCTCTGCCCCTGACGATGATACAGACATC-3’
TTS477-9AAA-S: 5’-CACTAAGAAAGCAGCCGCTGCTCTGGAATCATTCTTC-3’
TTS477-9DDD-S: 5’-CACTAAGAAAGCAGACGACGATCTGGAATCATTCTTC-3’
PCR reaction mixture (25 μl) contained reaction buffer supplied by the manufacturer, 0.25 mM dNTPs, 1 ng/μl plasmid, and indicated dose of primers. Primers used to introduce mutations to polη are shown above. The PCR products were digested by Dpn I (TaKaRa). Escherichia coli (E. coli) was transformed by digested DNA using the electroporation method, and transformants were seeded into the LB culture plate supplemented with 50 μg/ml carbenicillin to select a single colony. DNA was purified by the QIAprep Spin MiniPrep kit (QIAGEN), and mutations were confirmed by DNA sequence analysis. The plasmid DNA for transfection was prepared using the Endo-free® plasmid Maxi kit (QIAGEN).
Establishment of XP2SASV3 cells expressing mutants polη
Neon® transfection system (ThermoFisher) was used for plasmid transfection into cells according to the manufacture’s protocol. 10 μg of pIRESneo2 plasmids for expression of mutants polη were transfected into XP2SASV3 cells. Transfected cells were seeded into 100 mm dish cultured in Dulbecco’s modified Eagle’s medium (DMEM, WaKo) containing 10% fetal bovine serum (FBS, SIGMA). After 24 h incubation, cells were selected by 0.2 mg/ml G418 for two weeks.
Establishment of WI38VA13/Tet-off cells inducibly expressing polη
Tetracyclin-regulation pTRE-Tight plasmids for polη expression were linearized by PvuI (TaKaRa) before transfection. Linearized plasmid and 50 ng/μl linear hygr maker (TaKaRa) were transfected into WI38VA13/Tet-off cells (made by our group) using Neon® transfection system. Transfected cells were seeded into 60 mm dish and cultured in DMEM+10% FBS containing 100 ng/ml doxycycline. After 24 hours incubation, cells were reseeded into 100 mm dish and cultured in DMEM+10% FBS containing 0.1 mg/ml G418, 0.2 mg/ml hygromycin and 100 ng/ml doxycycline for 2 weeks. 500 cells of selected cells were seeded into 100 mm dish until forming colonies. Single colonies were transferred into 24 well plates. Cells were transferred to 6 well plates after growth in 24 well plates. At last, cells from 6 well plates were transferred to 100 mm dish.
Cell growth assay
3 × 105 cells were seeded into 60 mm culture dishes and cultured for 120 hours. Cells were harvested using trypsin and counted using Automated cell counter TC20TM (Bio-Rad) every 24 hours. Two dishes were used in each day. Graphs were written using the GraphPad Prism 8.
UV complementation assay
Cells were seeded in 6 well plates and cultured overnight. Cells were washed with 1X phosphate-buffered saline (PBS) and exposed to UV-C, then cultured cells in DMEM+10% FBS supplemented with 1 mM caffeine. After 4 days culture, cell viabilities were measure by MTS assay using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega).
Western blotting
Whole cell lysates were separated by SDS-PAGE and transferred to PVDF membranes (Millipore) using Trans-Blot SD Semi-Dry Transfer system (Bio-Red). After blocking with 5% skim milk in Tris-buffered saline containing tween-20 (TBS-T) (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% Tween 20), the membranes were incubated with primary antibodies, washed by TBS-T 4 times, every 5 minutes. Then the membranes were incubated with indicated secondary antibodies conjugated with horseradish peroxidase (HRP), and washed by TBS-T 4 times, every 5 minutes. Chem-Lumi One kit (Nacalai Tesque) was used for signal detection and Image QuantTM LAS4000 mini was used for visualization.
Results
Single consensus mutant of polη has equivalent effect with wild type polη
Human polη harbors three PIP boxes, whereas the amino acid sequences are non-canonical compared with the consensus sequence [24]. Three PIP boxes, especially PIP1 or PIP3, exhibit a weak interaction ability with PCNA [26], whereas the ability of the PIP box to interact with PCNA is a strong determinant of binding affinity [32]. Thus, we suspect that whether it’s promoting the affinity of PCNA when the PIP boxes in polη are converted into consensus sequence, the mutant polη increase the ability of UV resistance.
To investigate our hypothesis, we employed SV-40 transformed polη-deficient fibroblast XP2SASV3 cells and established XP2SASV3 cells stably expressing consensus mutants of polη. We introduced mutation at S437 residue of polη to glutamine and L444 to phenylalanine (S437Q/L444F, referred to as PIP1), which stably expressing single PIP1 consensus mutant. Subsequently, PIP2 consensus mutant (M701Q, referred to as PIP2), PIP3 consensus mutant (T477Q, referred to as PIP2) and wild type (WT) polη were also established (Figure 1A). As shown in Figure 1B, the expression level of WT and single consensus mutants of polη in XP2SASV3 cells were detected, whereas PIP1, PIP2, and PIP3 were almost similar but lower than WT.
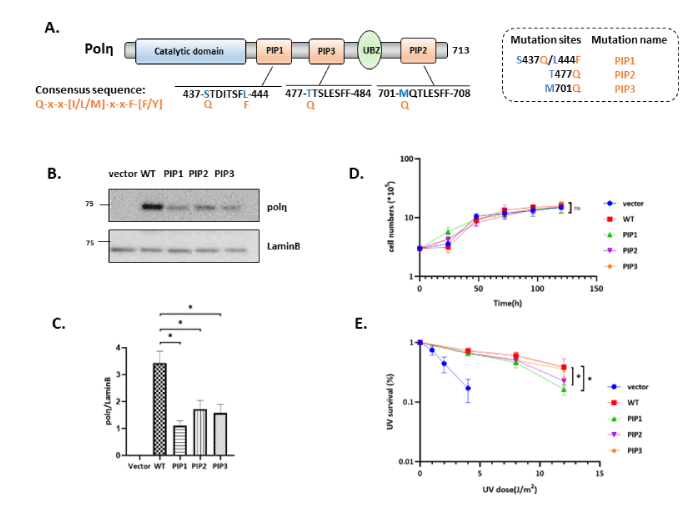
Figure 1: Single consensus mutant of polη has equivalent effect with wild type polη. (A) Schematic structure of human polη. PIP consensus sequence is also shown. Amino acid residues indicated by blue were changed to canonical amino acid Q or F. (B) Western blotting analysis of polη-expressing cells. Whole-cell lysates were prepared from XP2SASV3 cells expressing WT or single consensus mutants polη and analyzed by western blotting using anti-polη and anti-Lamin B. (C) Quantitative analysis of data from (B). (D) Growth curve of WT and mutants polη expressing cells. 3 × 105 cells were seeded, and cell numbers were counted every 24 h. The values represent the mean ± SD of two independent experiments. (E) Complementation activities of WT and mutants polη. Cells were irradiated with the indicated dose of UV-C and incubated with 1mM caffeine for 4 days. Viabilities of cells were measured by MTS assay. The values represent the mean ± SD of three independent experiments. *, P<0.05; ns: not significant.
We next asked whether the exogenous plasmid of consensus mutant affects the proliferation of XP2SASV3 cells. We observed almost similar growth rates among WT and PIP1, PIP3, and PIP2 consensus mutants polη expressing cells (Figure1D). Our UV sensitivity result showed that the PIP1 and PIP2 consensus mutant partially reduced the complementation activity of polη, whereas the PIP3 single mutant could complement the UV sensitivities of XP2SASV3 cells as same as WT polη (Figure1E). Thus, we conclude that single consensus mutant does not thoroughly alter WT polη function, suggesting that PIP3 consensus mutant likely performs equivalent effect with WT polη in XP2SASV3 cells. In contrast, PIP1 and PIP2 single consensus mutant might impact on polη routine lesion bypass ability.
Double consensus mutant of polη play alternative roles in cell proliferation
Since single consensus mutant of polη does not alter polη’s function much in response to UV irradiation, we hypothesized that expression of consensus mutant in double or triple PIP boxes might have the conspicuous effect (Figure 2A). To test this hypothesis, we tried to establish XP2SASV3 cells expressing PIP1/2 double consensus mutant of polη but haven’t succeeded. Thus, we suspected that the expression of double consensus mutant in XP2SASV3 cells might cause lethal defects and affect cell proliferation. To further examine the effect of expression of double or triple consensus mutants of polη, we considered employing a Tet-off inducible expression system that expresses proteins under doxycycline control. In Tet-off inducible expression system, when doxycycline was added to the cell culture medium (DOX+), expression of polη was suppressed. When doxycycline was eliminated from the cell culture medium (DOX-), expression of polη was induced (Figure 2B). The inducible expression plasmid for expression WT or double or triple consensus mutants of polη were transfected into SV-40 transformed normal human fibroblast WI38VA13/Tet-off cells and selected by 0.1 mg/ml G418 and 0.2 mg/ml hygromycin. Subsequently, we selected the single clones which can induce expression of WT polη or double or triple consensus mutants polη. Inducible expression of polη was detected by western blotting (Figure 2C). Indeed, the expression of WT and double consensus mutant polη up to the induction time. Under the above experimental conditions, inducible expressions of WT, PIP1/2, PIP1/3, PIP3/2, and PIP1/3/2 was detected to confirm successful cell construction (Figure 2E). To obtain further insight into the function of PIP1/2, PIP1/3, PIP3/2, and PIP1/3/2 mutants, we examined whether these mutants assuredly affect cell proliferation, as our speculation. Similarly, we first eliminated the doxycycline interference (Figure 3A) our results demonstrated that neither double consensus mutant nor triple consensus mutant could disturb the cell viability (Figure 3B-3G). In summary, these results indicate that double and triple consensus mutants show extraordinary pertinence to cell viability.
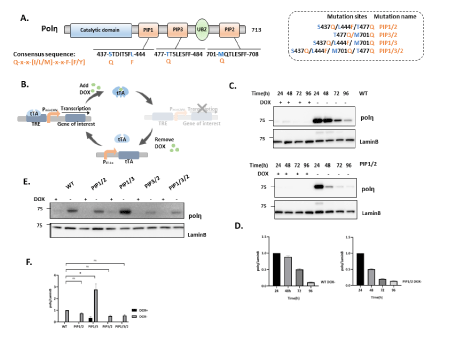
Figure 2: Double consensus mutant of polη plays alternative roles in cell proliferation. (A) Schematic structure of human polη. PIP consensus sequence is also shown. Amino acid residues indicated by blue were changed to canonical amino acid Q or F. (B) Schematic outline of Tet-OFF system with PCMV driving tTA expression. The binding of tTA protein to the TRE promoter is prevented by Dox administration (+Dox); withdrawal of Dox (-Dox) allows downstream effector gene transcription. (C) Time course analysis to confirm the optimal induce timepoint. Whole-cell lysates were prepared from WI38VA13/Tet-off cells expressing WT or PIP1/2 consensus mutants polη and analyzed by western blotting using anti-polη and anti-Lamin B. (D) Quantitative analysis of data from (C). (E) Western blotting analysis of polη-expressing cells. Whole-cell lysates were prepared from WI38VA13/Tet-off cells expressing WT, PIP1/2, PIP1/3, PIP3/2, and PIP1/3/2 consensus mutants polη and analyzed by western blotting using anti-polη and anti-Lamin B. (F) Quantitative analysis of data from (E) *, P<0.05; ns: not significant.
Double PIP boxes consensus mutant of Polη derivatives on tolerance to UV irradiation in WI38VA13/Tet-off cells
Considering that polη's significant functional role resides in its capability to bypass UV-induced CPD lesions, we asked whether the double and triple consensus mutant will have a more efficient ability to complement the UV sensitivity. We investigated whether the doxycycline affects polη's lesion bypassing ability. As we except, similar as proliferation assay, doxycycline also could not alter complementation activity of polη (Figure 4A-4E). Thus, we next estimated the lesion bypassing ability of inducible PIP boxes double and triple mutant polη without doxycycline, while there is no significant difference among all mutants (Figure 4F). These observations collectively allow us to speculate that PIP boxes double and triple consensus mutants of polη could take effect in perplexing ways. We considered that endogenous polη in WI38VA13 cells might have competitive impact on activities of inducibly expressed mutants polη.
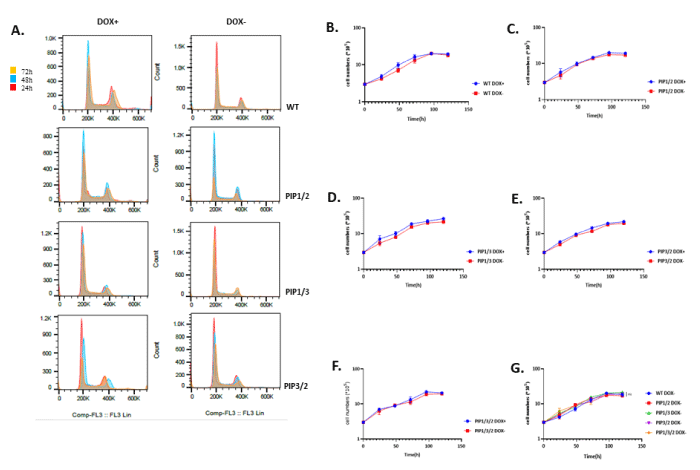
Figure 3: Double consensus mutant of polη plays alternative roles in cell proliferation. (A) Flow cytometry results show the cell cycle profile at various time intervals with or without doxycycline. (B-F) Growth curve of each cell cultured with or without doxycycline. 3 × 105 cells were seeded and cultured with or without doxycycline for 24 h, 1 × PBS and cultured in fresh medium for 24 h (Time 0 h). Cell numbers were counted every 24 h. The values represent the mean ± SD of two independent experiments. (G) Growth curve of WT polη and mutant polη expressing cells. 3 × 105 cells were seeded and cultured without doxycycline for 24 h, 1 × PBS and cultured in fresh medium for 24 h (Time 0 h). Cell numbers were counted every 24 h. The values represent the mean ± SD. of two independent experiments. ns: not significant.
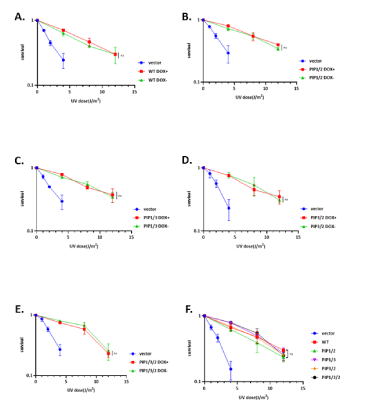
Figure 4: The role of double PIP boxes consensus mutant of polη in UV damage tolerance. (A-E) UV survival curves of WI38VA13/Tet-off cells stably expressing WT, double, and triple consensus mutant polη with or without doxycycline. Cells were irradiated with the indicated UV-C dose and incubated with 1mM caffeine for 4 days. Viabilities of cells were measured by MTS assay. (F) Complementation activities of WT and mutants polη without doxycycline. Cells were irradiated with the indicated UV-C dose and incubated with 1mM caffeine for 4 days. Viabilities of cells were measured by MTS assay. The values represent the mean ± SD of three independent experiments; ns: not significant.
Mutations in putative phosphorylation sites in/around PIP boxes of polη have an impact on the regulation of TLS mediated by polη after UV irradiation
Previous studies showed that phosphorylation of polη displayed lower survival after UV irradiation, indicating that phosphorylation of polη was involved in regulating UV damage bypassing [11,29-31]. We noticed peptides containing PIP1 and PIP3 of polη possess multiple serine and threonine amino acid which may act as potential phosphorylation sites. We subsequently employed a combined mass spectrometric and cell biological approach to identify and functionally characterize novel phosphorylation sites of polη in PIP boxes. As our hypothesis, PIP1 and PIP3 are phosphorylated in human cells (unpublished data). We employed deletion mutant carrying the 1-511 regions of polη (polηΔC) lacking PIP2, which can complement UV sensitivity of XP2SASV3 cells dependently on PIP1 and PIP3 [26]. The mass spectrometry assigned phosphorylation on S437 and T477, and other single phosphates on a peptide containing S435, SS436, S479. In this case, it was impossible to pinpoint exactly which of these sites impact the polη’s function. Thus, we first established the XP2SASV3 cells stably expressing phospho-mimetic PIP1 mutant S435D (SD), PIP3 mutant T477D (TD), and PIP1/PIP3 double mutant S437D/T477D (STD) of polηΔC (Figure 5A). A western blotting analysis compared mutant polηΔC levels in XP2SASV3 cells (Figure 5B). Like the previous report, our result also supported that polηΔC could almost complement the UV sensitivity of XP2SASV3 cells as WT did (Figure 5D). Subsequently, we examined the UV sensitivity of the above mutants. SD and TD mutations reduced complementation activity of polηΔC. STD mutation further reduced complementation ability compared with SD and TD single mutation, suggesting that the S/T to D mutations inhibit PIP1 and PIP3 function. In this vein, it is worth noting that there are multiple S and T around PIP1 S437 and PIP3 T477 sites. Thus, we next constructed XP2SASV3 cells stably expressing phosphor-mimetic mutants SSS435-7DDD (SSSDDD), TTS477-9DDD (TTSDDDD) and SSS435-7DDD/TTS477-9DDD(SDTD) of polηΔC, and phospho-deficient mutants SSS435-7AAA (SSSAAA), TTS477-9AAA (TTSAAA), SSS435-7AAA/TTS477-9AAA (SATA) of polηΔC (Figure 6A). We confirmed that the expression levels of mutant polη were similar in the transfected cells (Figure 6B, 6E). As shown in Figure 6G, sensitivities of XP2SASV3 cells after UV irradiation were restored to a similar extent by polηΔC, SSSAAA, TTASAAA, and SATA polηΔC mutant. SSSDDD, TTSDDD mutations reduced complementation activity of polηΔC. SDTD mutation further reduced complementation ability compared with single mutation, suggesting that the S/T to D mutations inhibit PIP1 and PIP3 function as Figure 4F showed (Figure 6D). Together, the information implies that phosphorylation of polη in PIP box might negatively regulate the polη UV-induced lesion bypassing ability.
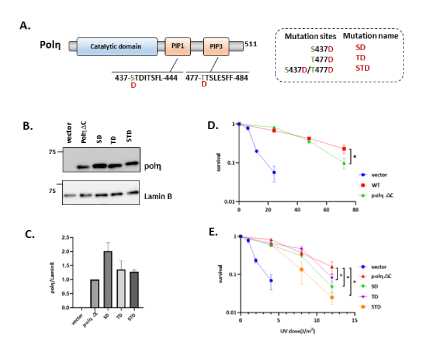
Figure 5: The role of S437 and T477 phosphorylation in UV damage tolerance. (A) Schematic structure of human polηΔC. Amino acid residues indicated by green were replaced with D. Amino acid residues indicated by the underline show PIP1 and PIP3. (B) Western blotting analysis of polηΔC-expressing cells. Whole-cell lysates were prepared and analyzed by western blotting using anti-polη and anti-Lamin B. (C) Quantitative data analysis from (B). (D,E) UV complementation assay of XP2SASV3 cells stably expressing WT polηΔC and mutant polηΔC. Cells were irradiated with the indicated UV-C dose and incubated with 1 mM caffeine for 4 days. Viabilities of cells were measured by MTS assay. The values represent the mean ± SD of three independent experiments. *, P<0.05; ns: not significant.
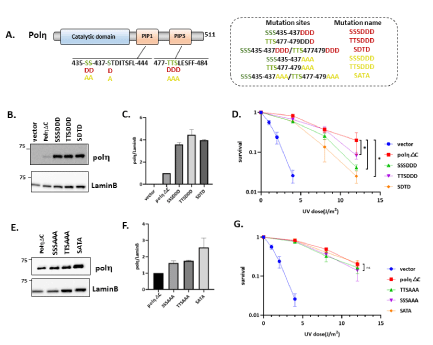
Figure 6: Mutations in putative phosphorylation sites in/around PIP boxes of polη affect the regulation of TLS mediated by polη after UV irradiation. (A) Schematic structure of human polηΔC. Amino acid residues indicated by green were replaced with D or A. Amino acid residues indicated by the underline show PIP1 and PIP3. (B,E) Western blotting analysis of polηΔC-expressing cells. Whole cell lysates were prepared and analyzed by western blotting using anti-polη and anti-Lamin B. (C,F) Quantitative analysis of data from (B,E). (D,G) UV complementation assay of XP2SASV3 cells stably expressing WT polηΔC and mutant polηΔC. Cells were irradiated with indicated dose of UV-C, and incubated with 1 mM caffeine for 4 days. Viabilities of cells were measured by MTS assay. The values represent the mean ± SD of three independent experiments. *, P<0.05; ns: not significant.
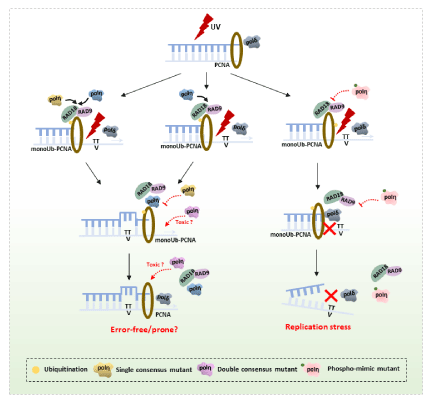
Figure 7: Proposed working model for mutant polη in response to UV irradiation in mammalian cells. UV irradiation- induced DNA damage blocks replicative polymerase like Polδ. The ssDNA and stalled replication fork recruit the Rad6-Rad18 complex to monoubiquitinate PCNA-K164, which in turn recruits polη. Single consensus mutant competing with WT polη has the equivalent effect with polη to perform error-free DNA damage tolerance. However, double consensus mutant polη in XP2SASV3 cells have serve cytotoxicity, while endogenous polη in WI38VA13 cells may have competitive effects on ability of inducibly expressed mutants polη. In addition, phosphorylation of polη in PIP1 and PIP3 negatively regulates polη lesion bypassing ability resulting in replication stress.
Discussion
Genetic deficiency in polη causes xeroderma pigmentosum variant syndrome in humans, which is manifested by sunlight sensitivity and elevated susceptibility to developing sunlight-induced skin cancer [33]. DNA polymerase η bypasses CPDs via an error-free manner, protects the genome from DNA replication stress [34,35]. Regardless of this characteristic, polη is potentially an error-prone polymerase along with the other TLS polymerases when encounter undamaged DNA [36]. Thus, the polymerase switching process in TLS should be strictly regulated. The PIP boxes of polη execute protein-protein interactions and intracellular localization in response to UV damage [37].
In this study, we used the polη-deficient XP2SASV3 cell expressing the PIP1, PIP3, and PIP2 single consensus mutants of polη to verify the function of consensus PIP boxes mutants, whereas no significant differences were observed among these cells. We also observed that either PIP1, PIP3, or PIP2 consensus mutants of polη could complement UV sensitivities of XP2SASV3 cells as WT polη. Based on these findings, we proposed that single consensus PIP mutants polη may have equivalent abilities to WT polη. However, the expression of PIP1/2 double consensus mutant drastically affects proliferation of XP2SASV3 cells, which supporting that double consensus mutant may have severe cytotoxicity. Thereby, we employed the Tet-off inducible expression system in WI38VA13 cells to examine the effects of expression of double and triple consensus mutants, astonishingly, no dramatical discrepancy among these cells in growth rate and UV damage bypassing ability. We speculated that endogenous polη in WI38VA13 cells might have competitive effects on activities of inducibly expressed mutants polη. It is worth exploring that double and triple consensus mutants could take effect in mysterious ways. Honestly, the current results are insufficient to confirm the above conclusion; more experiments are needed to further explore the detailed mechanisms.
In human cells, the current working model is that TLS polymerases are controlled by modulating their access to the replication fork by PTMs [11,27,38]. In the case of polη, phosphorylation of multiple sites in polη is involved in TLS regulations. In this study, we examined the possibility that phosphorylation of PIP1 and PIP3 is involved in the regulation of polη. Replacement of S and/or T residues in PIP1 or PIP3 to D reduced the complementation activity of polηΔC, whereas A did not. The S/T to D mutations in PIP1 and PIP3 of polηΔC further reduced the complementation ability compared to the single mutants. In contrast, the S/T to A mutant complemented the UV sensitivity of XP2SASV3 cells as polηΔC. Previous studies showed that phosphorylation of polη positively regulates TLS [11,29-31], in contrast, our results suggested that phosphorylations of polη in its PIP1 or PIP3 might negatively regulate polη’s function, which may disturb the polymerase switch resulting in the replication stress. We have so far not been able to detect the kinase of these phosphorylation directly. We therefore would like to investigate if these phosphorylation’s are a specific kinase target through an alternative approach. Taken together, we present a speculative model in Figure 7 that is consistent with our data.
In summary, we demonstrated that PIP1, PIP2, and PIP3 are crucial modules to regulate polη’s function. We revealed that PIP boxes consensus mutant polη has a dual role in performing its functions and in polymerase switching, where consensus mutant polη compete with wild type polη to execute lesion bypass. In contrast, we identified PIP1 and PIP3 as novel phosphorylation sites of human polη by mass spectrometric analysis. Moreover, phosphorylation’s of polη in its PIP1 and PIP3 negatively regulate polη’s function, supporting that the post-translational modifications of the PIP boxes of polη functions in UV damage bypassing.
Acknowledgements
We would like to thank Rie Kanao, Chikahide Masutani, and all the members of the lab in Nagoya University for their support.
Conflict of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This work was financially supported by Fundamental Research Funds for the Central Universities (lzujbky-2022-it14).
References
- Maya-Mendoza A, Moudry P, Merchut-Maya JM, Lee M, Strauss R, et al. (2018) High speed of fork progression induces DNA replication stress and genomic instability. Nature 559: 279-284. [Crossref]
- Ghosh S, Ghosh A (2021) Activation of DNA damage response signaling in mammalian cells by ionizing radiation. Free Radic Res 55: 581-594. [Crossref]
- Kang TH (2021) Circadian Rhythm of NER and ATR Pathways. Biomolecules 11: 715. [Crossref]
- Long LJ, Lee PH, Small EM, Hillyer C, Guo Y, et al. (2020) Regulation of UV damage repair in quiescent yeast cells. DNA Repair (Amst) 90: 102861. [Crossref]
- Branzei D, Psakhye I (2016) DNA damage tolerance. Curr Opin Cell Biol 40: 137-144. [Crossref]
- Kanao R, Masutani C (2017) Regulation of DNA damage tolerance in mammalian cells by post-translational modifications of PCNA. Mutat Res 803-805: 82-88. [Crossref]
- Maiorano D, El Etri J, Franchet C, Hoffmann JS (2021) Translesion Synthesis or Repair by Specialized DNA Polymerases Limits Excessive Genomic Instability upon Replication Stress. Int J Mol Sci 22: 3924. [Crossref]
- Kusumoto R, Masutani C, Iwai S, Hanaoka F (2002) Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry 41: 6090-6099. [Crossref]
- Kanao R, Yokoi M, Ohkumo T, Sakurai Y, Dotsu K, et al. (2015) UV-induced mutations in epidermal cells of mice defective in DNA polymerase η and/or ι. DNA Repair (Amst) 29: 139-146. [Crossref]
- Acharya N, Manohar K, Peroumal D, Khandagale P, Patel SK, et al. (2019) Multifaceted activities of DNA polymerase η: beyond translesion DNA synthesis. Curr Genet 65: 649-656. [Crossref]
- Bertoletti F, Cea V, Liang CC, Lanati T, Maffia A, et al. (2017) Phosphorylation regulates human polη stability and damage bypass throughout the cell cycle. Nucleic Acids Res 45: 9441-9454. [Crossref]
- González-Magaña A, Blanco FJ (2020) Human PCNA Structure, Function and Interactions. Biomolecules 10: 570. [Crossref]
- Boehm EM, Gildenberg MS, Washington MT (2016) The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes 39: 231-254. [Crossref]
- Acharya N, Patel SK, Sahu SR, Kumari P (2020) 'PIPs' in DNA polymerase: PCNA interaction affairs. Biochem Soc Trans 48: 2811-2822. [Crossref]
- Lee KY, Park SH (2020) Eukaryotic clamp loaders and unloaders in the maintenance of genome stability. Exp Mol Med 52: 1948-1958. [Crossref]
- Shen S, Davidson GA, Yang K, Zhuang Z (2021) Photo-activatable Ub-PCNA probes reveal new structural features of the Saccharomyces cerevisiae Polη/PCNA complex. Nucleic Acids Res 49: 9374-9388. [Crossref]
- Acharya N, Yoon JH, Gali H, Unk I, Haracska L, et al. (2008) Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc Natl Acad Sci U S A 105: 17724-17729. [Crossref]
- Che J, Hong X, Rao H (2021) PCNA Ubiquitylation: Instructive or Permissive to DNA Damage Tolerance Pathways?. Biomolecules 11: 1543. [Crossref]
- Pastushok L, Hanna M, Xiao W (2010) Constitutive fusion of ubiquitin to PCNA provides DNA damage tolerance independent of translesion polymerase activities. Nucleic Acids Res 38: 5047-5058. [Crossref]
- Lau WC, Li Y, Zhang Q, Huen MS (2015) Molecular architecture of the Ub-PCNA/Pol η complex bound to DNA. Sci Rep 5: 15759. [Crossref]
- Krijger PH, van den Berk PC, Wit N, Langerak P, Jansen JG, et al. (2011) PCNA ubiquitination-independent activation of polymerase η during somatic hypermutation and DNA damage tolerance. DNA Repair (Amst) 10: 1051-1059. [Crossref]
- Prestel A, Wichmann N, Martins JM, Marabini R, Kassem N, et al. (2019) The PCNA interaction motifs revisited: thinking outside the PIP-box. Cell Mol Life Sci 76: 4923-4943. [Crossref]
- Horsfall AJ, Vandborg BA, Kowalczyk W, Chav T, Scanlon DB, et al. (2021) Unlocking the PIP-box: A peptide library reveals interactions that drive high-affinity binding to human PCNA. J Biol Chem 296: 100773. [Crossref]
- Hishiki A, Hashimoto H, Hanafusa T, Kamei K, Ohashi E, et al. (2009) Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J Biol Chem 284: 10552-10560. [Crossref]
- Biertümpfel C, Zhao Y, Kondo Y, Ramón-Maiques S, Gregory M, et al. (2010) Structure and mechanism of human DNA polymerase eta. Nature 465: 1044-1048. [Crossref]
- Masuda Y, Kanao R, Kaji K, Ohmori H, Hanaoka F, et al. (2015) Different types of interaction between PCNA and PIP boxes contribute to distinct cellular functions of Y-family DNA polymerases. Nucleic Acids Res 43: 7898-7910. [Crossref]
- Wu PS, Enervald E, Joelsson A, Palmberg C, Rutishauser D, et al. (2020) Post-translational Regulation of DNA Polymerase η, a Connection to Damage-Induced Cohesion in Saccharomyces cerevisiae. Genetics 216: 1009-1022. [Crossref]
- Chen YW, Cleaver JE, Hatahet Z, Honkanen RE, Chang JY, et al. (2008) Human DNA polymerase eta activity and translocation is regulated by phosphorylation. Proc Natl Acad Sci U S A 105: 16578-16583. [Crossref]
- Dai X, You C, Wang Y (2016) The Functions of Serine 687 Phosphorylation of Human DNA Polymerase η in UV Damage Tolerance. Mol Cell Proteomics 15: 1913-1920. [Crossref]
- Göhler T, Sabbioneda S, Green CM, Lehmann AR (2011) ATR-mediated phosphorylation of DNA polymerase η is needed for efficient recovery from UV damage. J Cell Biol 192: 219-227. [Crossref]
- Peddu C, Zhang S, Zhao H, Wong A, Lee EYC, et al. (2018) Phosphorylation Alters the Properties of Pol η: Implications for Translesion Synthesis. iScience 6: 52-67. [Crossref]
- Mailand N, Gibbs-Seymour I, Bekker-Jensen S (2013) Regulation of PCNA-protein interactions for genome stability. Nat Rev Mol Cell Biol 14: 269-282. [Crossref]
- Yang Y, Gao Y, Zlatanou A, Tateishi S, Yurchenko V, et al. (2018) Diverse roles of RAD18 and Y-family DNA polymerases in tumorigenesis. Cell Cycle 17: 833-843. [Crossref]
- Hendel A, Ziv O, Gueranger Q, Geacintov N, Livneh Z (2008) Reduced efficiency and increased mutagenicity of translesion DNA synthesis across a TT cyclobutane pyrimidine dimer, but not a TT 6-4 photoproduct, in human cells lacking DNA polymerase eta. DNA Repair (Amst) 7: 1636-1646. [Crossref]
- Quinet A, Martins DJ, Vessoni AT, Biard D, Sarasin A, et al. (2016) Translesion synthesis mechanisms depend on the nature of DNA damage in UV-irradiated human cells. Nucleic Acids Res 44: 5717-5731. [Crossref]
- Vaziri C, Rogozin IB, Gu Q, Wu D, Day TA (2021) Unravelling roles of error-prone DNA polymerases in shaping cancer genomes. Oncogene 40: 6549-6565. [Crossref]
- Tsanov N, Kermi C, Coulombe P, Van der Laan S, Hodroj D, et al. (2014) PIP degron proteins, substrates of CRL4Cdt2, and not PIP boxes, interfere with DNA polymerase η and κ focus formation on UV damage. Nucleic Acids Res 42: 3692-3706. [Crossref]
- Guérillon C, Smedegaard S, Hendriks IA, Nielsen ML, Mailand N (2020) Multisite SUMOylation restrains DNA polymerase η interactions with DNA damage sites. J Biol Chem 295: 8350-8362. [Crossref]







