In order to identify proteins differentially expressed in rheumatoid arthritis patients and healthy controls, we used isobaric tagging for relative and absolute protein quantification (iTRAQ) to generate quantitative protein profiles from peripheral blood mononuclear cells (PBMCs) from 10 patients with rheumatoid arthritis and 10 healthy donors. Proteins were extracted, concentrated, pooled, labeled with iTRAQ reagents and subjected to multi-dimensional chromatographic fractionation followed by tandem mass spectrometry. A total of 3,208 proteins were identified based on ProteinPilotTM 3.0 software and the International Protein Index (human 3.62) database. Of these, 83 proteins differed at least 2-fold in expression levels between patients and controls. These results indicate that iTRAQ-based technology can serve as a useful aid for identification and quantification of disease-linked proteins from PBMCs. Several proteins, such as serotransferrin, clusterin, annexin A2, and heterogeneous nuclear ribonucleoprotein A2/B1, may be served as biomarker candidates.
iTRAQ, Proteomics, Rheumatoid Arthritis
Rheumatoid arthritis is an autoimmune inflammatory rheumatic disease that affects many tissues and organs and causes chronic synovial inflammation, eventually leading to joint destruction and disability [1-4]. The pathogenesis of rheumatoid arthritis is quite complex and involves genetic and environmental factors. One of the main goals of research on this disease is to identify biomarkers that can shed light on disease onset and progression as well as improve diagnosis, treatment and prognosis.
Several studies implicate lymphocytes in the pathology of rheumatoid arthritis [5-8]. Therefore, we wanted to focus on peripheral blood mononuclear cells (PBMCs) and compare their proteomic profiles from patients and healthy controls. To avoid the problems of high background and low sensitivity associated with two-dimensional electrophoresis and other proteomic approaches used previously [9-11] to examine PBMCs in patients with rheumatoid arthritis, we chose the method of isobaric tagging for relative and absolute quantitation (iTRAQ). This has proven to be a powerful new tool for proteomic research in many autoimmune diseases [10,12-16]. Up to eight tags can be used in multiplex iTRAQ [17,18], and the large amounts of data emerging from tandem mass spectrometry analysis can be analyzed using bioinformatics. Despite its efficiency as a protein discovery tool, iTRAQ has yet to be applied to rheumatoid arthritis based on our review of the literature.
Here we used iTRAQ-based quantitative proteomic analysis and bioinformatics-based protein identification in order to identify proteins linked to rheumatoid arthritis. We identified these proteins by comparing proteomic profiles of PBMCs from patients and healthy controls.
Patients
PBMCs were isolated from 10 patients with rheumatoid arthritis (five men; mean age, 45 yr; age range, 36 to 56 yr. five women; mean age, 47 yr; age range, 39 to 55 yr) who fulfilled the American College of Rheumatology Revised Criteria for Rheumatoid Arthritis, as well as from 10 healthy controls who were age- and sex-matched with the patients. Patients had no history of other immune diseases, such as ankylosing spondylitis, systemic lupus erythematosus, or multiple sclerosis. Patients had not received any drug treatment prior to blood sample collection.
All participants provided written informed consent for this study, which was performed at Xinyang Normal University and Xinyang First People's Hospital (Railway Hospital) in China. The study protocol was approved by the institutional research ethics committees at both study sites.
PBMC isolation and protein extraction
Venous blood (4 mL) was collected from all patients and controls into EDTA anticoagulant tubes. PBMC were isolated using lymphocyte separation medium following the manufacturer’s instructions (Cedarlane Laboratories, Hornby, Ontario, Canada). Total protein was extracted from PBMCs, and concentration was measured using the BCA protein assay kit (Pierce Biotechnology, Rockford, IL, USA). Pools of 100 μg of total protein from patients and controls [19] were prepared and analyzed using iTRAQ, respectively.
iTRAQ proteomics
Protein (100 μg) from two patient pools and two control pools was blocked, digested, and labeled according to the iTRAQ protocol (Applied Biosystems, Foster City, CA, USA). Four iTRAQ tagging reactions were prepared (Patient Pool 1-iTRAQ 117, Patient Pool 2-iTRAQ 118, Control Pool 1-iTRAQ 119 and Control Pool 2-iTRAQ 121), then all reactions were pooled into a single mixture, which was fractionated by multi-dimensional liquid chromatography involving a Zorbax Bio-SCX column (35×0.3 mm, 300 Å, 3.5 μm particles; Agilent Technologies, Santa Clara, CA, USA) and a gradient of potassium formate in 25% acetonitrile, which generated 10 strong cation exchange fractions; these fractions were further separated on a TempoTM LC nanoflow and MALDI spotting system (Applied Biosystems) equipped with a reverse-phase Magic C18AQ column (150×0.2 mm, 200 Å , 3 μm particles; Michrom Bioresources, CA, USA). Each multidimensional chromatography run yielded approximately 380 MALDI spots on a stainless steel MALDI target plate.
Mass spectrometry
Mass spectrometry was performed on an Applied Biosystems 4800 Plus MALDI TOF/TOF system. Only results associated with a signal-to-noise ratio of at least 40 were selected for tandem mass spectrometry. Mass spectra from 500 laser shots were acquired for each spot. The tandem mass spectrometry data from all 10 fractions were input into a Paragon algorithm search conducted using Protein PilotTM 3.0 software (Applied Biosystems/MDS-Sciex) against a database of the International Protein Index (human, version 3.62) [20], which was downloaded from the EBI website (www.ebi.ac.Uk/IPI/IPIhelp.html). Only proteins identified with at least 95% confidence, or a ProtScore of 1.3, were analyzed in detail. Relative quantification of proteins was based on the ratio of peak areas of m/z 117, 118, 119, and 121 from tandem mass spectrometry spectra.
We used the following procedure to define the minimal difference in relative levels of a protein between patients and controls that would allow us to define a protein as linked to rheumatoid arthritis. Two equal amounts of 10-fraction mixtures (Applied Biosystems) were trypsin-digested and labeled with iTRAQ reagents. The standard deviation (SD) of all the ratios of the labeled peptides was computed to be 0.15. This led to a cut-off value of 2 SD + 1 = 1.3 as the minimal fold-change in patients relative to controls necessary for defining a protein as up-regulated in disease. The reciprocal cut-off of 0.77 was used as the maximal fold-change in patients relative to controls necessary for defining a protein as down-regulated in disease. These same cut-offs were also used to filter out proteins that differed by more than 1.3-fold or less than 0.77-fold between the two patient pools and the two control pools (technical replicates).
Bioinformatics analysis
Candidate biomarkers of rheumatoid arthritis were functionally annotated based on the Gene Ontology (GO) classification system (http://wego.genomics.org.cn), and a network of interacting molecules was built using on-line Ingenuity Pathway Analysis (IPA, www.ingenuity.com).
Identification of protein expression changes by iTRAQ proteomic analysis
A total of 3,208 proteins were identified from patient and control PBMC extracts, of which 42 were significantly up-regulated in rheumatoid arthritis and 41 were down-regulated (Table 1). Figure 1 shows a representative tandem mass spectrum of serotransferrin, one of the proteins up-regulated in disease. Figure 2 shows the spectrum of annexin A2, one of the proteins down-regulated in disease.
Table 1. Proteins showing the greatest fold-changes in patients with rheumatoid arthritis relative to healthy controls.
No. |
Accession code |
Name |
Average folds (arthritis/control) |
Top 10 proteins up-regulated in rheumatoid arthritis |
1 |
sp|P01859 |
Ig gamma-2 chain C region |
4.823439138 |
2 |
sp|P02768 |
Serum albumin |
4.773611854 |
3 |
sp|P01834 |
Ig kappa chain C region |
4.750356564 |
4 |
sp|P02787 |
Serotransferrin |
4.667309404 |
5 |
sp|P00738 |
Haptoglobin |
4.089293455 |
6 |
sp|P01009 |
Alpha-1-antitrypsin |
3.903263238 |
7 |
sp|P01857 |
Ig gamma-1 chain C region |
3.803608909 |
8 |
sp|P01615 |
Immunoglobulin kappa variable 2D-28 |
3.693985617 |
9 |
sp|P01876 |
Ig alpha-1 chain C region |
3.424910075 |
10 |
sp|P02671 |
Fibrinogen alpha chain |
3.371755769 |
Top 10 proteins down-regulated in rheumatoid arthritis |
1 |
sp|Q16777 |
Histone H2A type 2-C |
4.24889502 |
2 |
sp|P16402 |
Histone H1.3 |
4.101023024 |
3 |
sp|P41218 |
Myeloid cell nuclear differentiation antigen |
3.913870339 |
4 |
sp|Q96L21 |
60S ribosomal protein L10-like |
3.653741806 |
5 |
sp|P18859 |
ATP synthase-coupling factor 6, mitochondrial |
3.34532721 |
6 |
sp|P62266 |
40S ribosomal protein S23 |
3.209125754 |
7 |
sp|P06454 |
Prothymosin alpha |
3.192761477 |
8 |
sp|P61769 |
Beta-2-microglobulin |
3.176176391 |
9 |
sp|Q07020 |
60S ribosomal protein L18 |
3.155362286 |
10 |
sp|Q9H299 |
SH3 domain-binding glutamic acid-rich-like protein 3 |
3.122316334 |
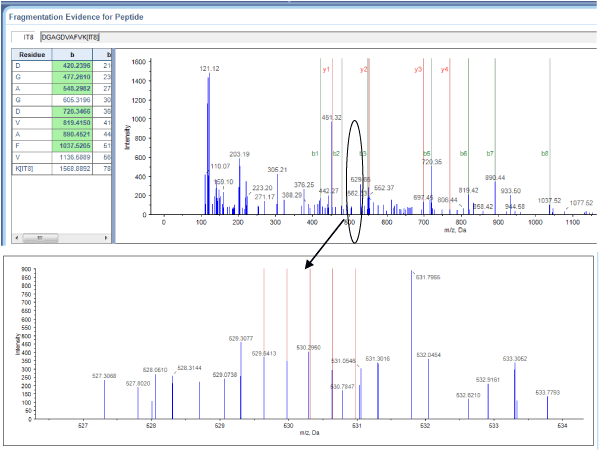
Figure 1. Serotransferrin sequencing and quantification using iTRAQ. A representative MALDI TOF/TOF MS/MS spectrum of the serotransferrin-derived peptide DGAGDVAFVK is shown with fragmented b-ions and y-ions.
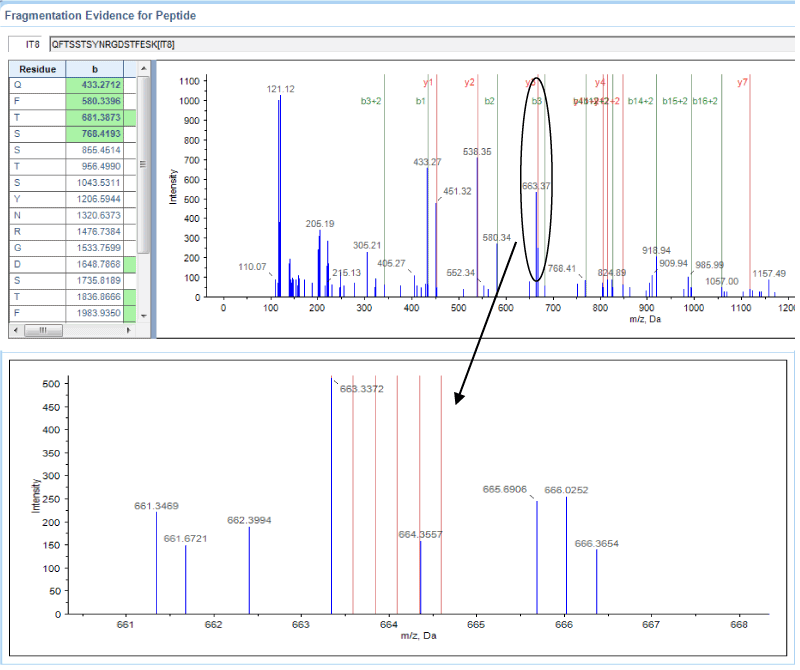
Figure 2. Annexin A2 sequencing and quantification using iTRAQ. A representative MALDI TOF/TOF MS/MS spectrum of the Annexin A2-derived peptide QFTSSTSYNRGDSTFESK is shown with fragmented b-ions and y-ions.
Potential functions of proteins linked to rheumatoid arthritis
GO analyses using the WEGO online tool indicated that the proteins significantly up- and down-regulated in patients relative to controls are involved in cellular and metabolic processes, biological regulation, and molecular functions of binding, catalysis and transport (Figure 3).
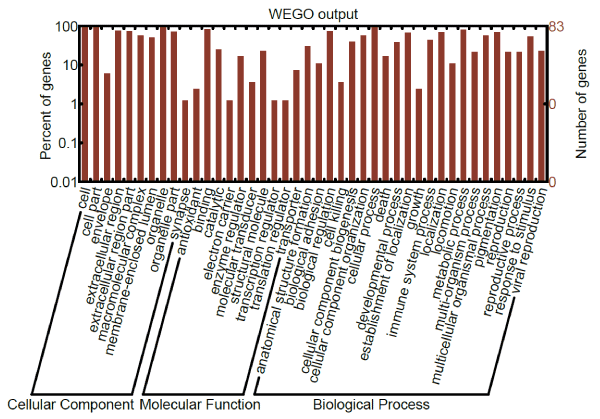
Figure 3. GO analysis of proteins significantly up- or down-regulated in rheumatoid arthritis.
Pathway and networks involving proteins linked to rheumatoid arthritis
IPA was used to construct pathways and networks involving proteins significantly up- or down-regulated in rheumatoid arthritis in order to analyze their possible relationships (Figure 4). The pathways are focused on receptor signal activation and acute phase response signaling, while the six networks involve primarily organismal injury and abnormalities, immunological disease, cellular movement, connective tissue disorders, cell death and survival, post-translational modification, and cell-to-cell signaling and interaction (Figure 5).
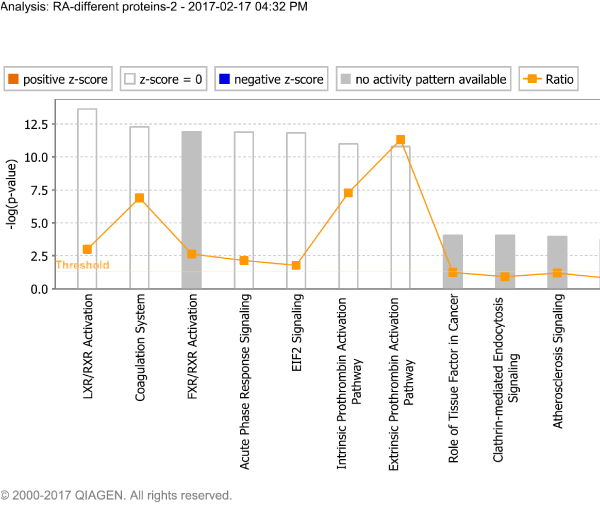
Figure 4. Signaling pathways involving proteins significantly up- or down-regulated in rheumatoid arthritis.
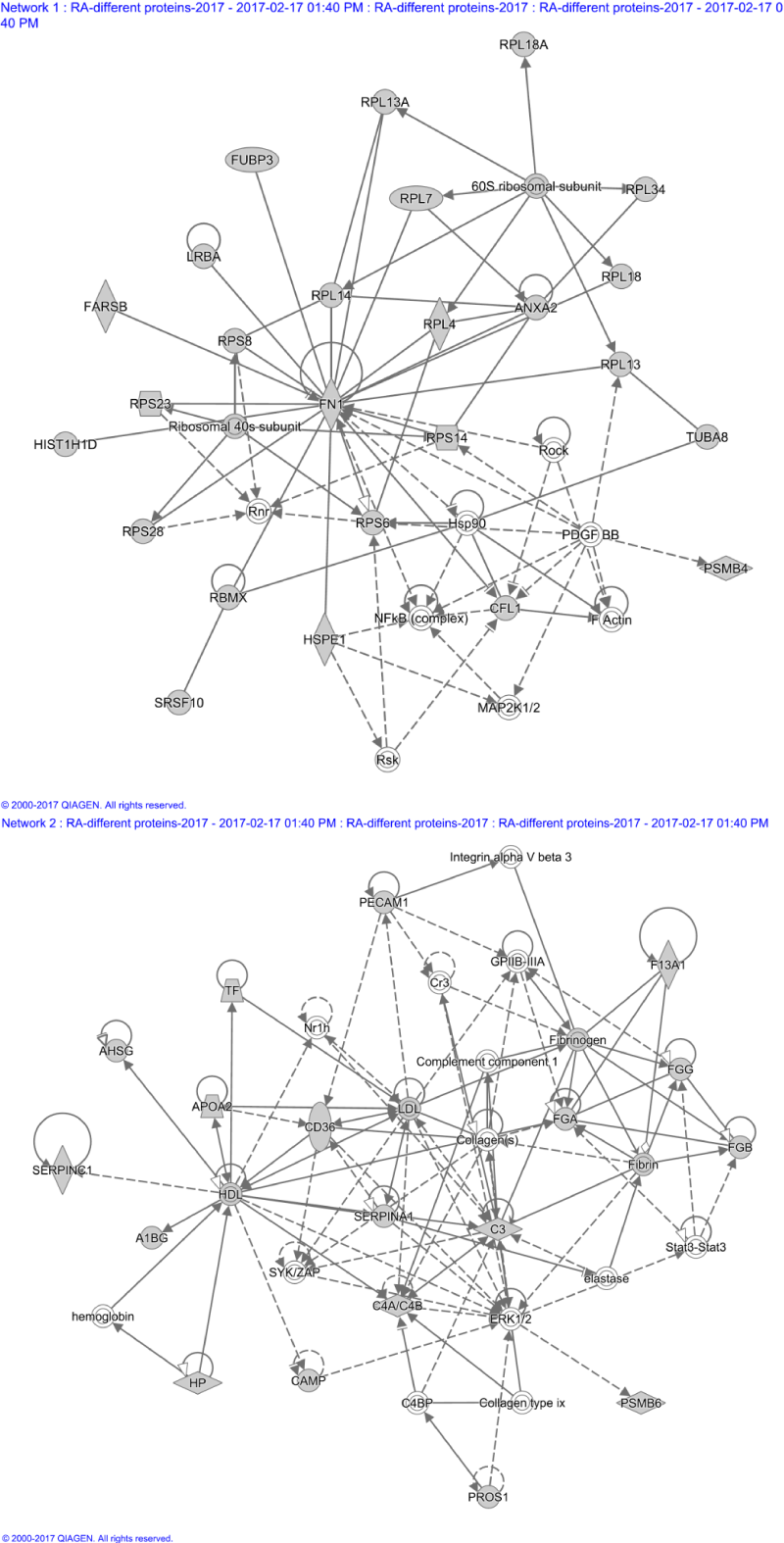
Figure 5. Two networks involving proteins significantly up- or down-regulated in rheumatoid arthritis.
(A) A network involved in organismal injury and abnormalities. (B) A network involved in organismal functions, immunological disease, and cellular movement.
Little is known about proteins associated with the pathogenesis of rheumatoid arthritis, and several are likely to play key roles, given that the disease is likely to involve many genes. Here we identified several proteins significantly up- and down-regulated in the disease, providing the basis for further molecular studies aimed at identifying biomarkers useful for diagnosis, treatment and prognosis. While iTRAQ has proven effective at identifying disease biomarkers from tissue or serum [9-11,15,19,21-23], its usefulness for analyzing proteins from PBMCs is only beginning to be explored in the case of rheumatoid arthritis.
In this study, 83 proteins were identified as differentially expressed in rheumatoid arthritis. Most are inflammation-associated proteins and many were found to be up-regulated in the disease, including complement, immunoglobulin, and ribosomal protein. These proteins seem less useful as potential rheumatoid arthritis biomarkers, since they are also up-regulated in other auto-immune diseases. In contrast, other proteins that we identified as linked to rheumatoid arthritis merit further attention as biomarker candidates, including serotransferrin, clusterin, annexin A2, and heterogeneous nuclear ribonucleoprotein A2/B1.
GO analysis suggested that many of the proteins linked to rheumatoid arthritis in our study are involved in cellular and metabolic processes, biological regulation, and molecular functions related to binding, catalysis and transport. Interaction networks constructed from all 83 disease-linked proteins are involved primarily in organismal injury and abnormalities, immunological disease, cellular movement, connective tissue disorders, cell death and survival, post-translational modification, and cell-to-cell signaling and interaction.
Several proteins identified in our study as linked to rheumatoid arthritis have already been shown in previous work to contribute to pathogenesis of the disease. Clusterin, which was up-regulated in disease in our study, is expressed predominantly by synoviocytes and is present in synovial fluid. It promotes apoptosis and can regulate the NF-kappaB pathway, strongly affecting inflammation and immunity [24,25]. Annexin A3, which was up-regulated in disease in our study, induces migration and tube formation of human umbilical vein endothelial cells; transfecting cells with an annexin A3 expression plasmid leads to high levels of vascular endothelial growth factor (VEGF), a major angiogenic factor. Annexin A3 also enhances the transactivation activity of hypoxia-inducible factor-1 (HIF-1) [26]. The consistency between these previous studies and our own findings suggests that our iTRAQ results are reliable.
Some of the proteins down-regulated in rheumatoid arthritis in our study have previously been shown to be down-regulated in other disease contexts. Prothymosin α exerts in vitro immunomodulatory effects on autoimmune diseases [27-29]. Cytochrome c oxidase (CytcO) is a multi-subunit enzyme that acts as both an O2 sensor and a regulatory enzyme that determines mitochondrial respiration capacity. Hypoxia inhibits CytcO expression in vitro [30], which is linked with joint inflammation and decreased vascularity [31,32]. Positively charged cytochrome c and negatively charged truncated prothymosin α, when meeting in cytosol, can interact with each other. Truncated prothymosin α inhibits cytochrome c oxidation by H2O2, catalyzed by peroxidase [33]. Down-regulation of CytcO has been reported in juvenile idiopathic arthritis [34] and inflammatory arthritis.
Our iTRAQ study identified some proteins, such as beta-2-microglobulin, thymosin beta4 and beta10, have not previously been linked to rheumatoid arthritis, but have been linked to other diseases. Examples are beta-2-microglobulin and thymosin β10 [35-38]. These candidate rheumatoid arthritis biomarkers deserve further attention.
The goal of the present study was to identify a set of potential candidate biomarkers of rheumatoid arthritis that could be validated and further analyzed in future work involving other patient populations. Such work should verify and extend the present results by comparing protein levels in PBMC and serum between patients and controls.
2021 Copyright OAT. All rights reserv
The authors declare no conflicts of interest.
Supported by Henan science and technology innovation team, Investigation on plant resources in Dabie Mountains and the study and utilization of active components of special plants, Nanhu Scholars Program for Young Scholars of XYNU
- Gilkeson GS, Allen NB (1996) Retroperitoneal fibrosis. A true connective tissue disease. Rheum Dis Clin North Am 22: 23-38. [Crossref]
- Meier P, Gilabert C, Burnier M, Blanc E (2003) La fibrose rétropéritonéale, une maladie inflammatoire méconnue. Observations cliniques et revue de la littérature. Nephrologie 24: 173-180.
- Vaglio A, Salvarani C, Buzio C (2006) Retroperitoneal fibrosis. Lancet 367: 241-251. [Crossref]
- Vaglio A, Corradi D, Manenti L, Ferretti S, Garini G, et al. (2003) Evidence of autoimmunity in chronic periaortitis: a prospective study. Am J Med 114: 454-462. [Crossref]
- Van Bommel EF, Jansen I, Hendriksz TR, Aarnoudse AL (2009) Idiopathic retroperitoneal fibrosis: prospective evaluation of incidence and clinicoradiologic presentation. Medicine (Baltimore) 88: 193-201.
- Vaglio A, Palmisano A, Alberici F, Maggiore U, Ferretti S, et al. (2011) Prednisone versus tamoxifen in patients with idiopathic retroperitoneal fibrosis: an open-label randomised controlled trial. Lancet 378: 338-346. [Crossref]
- Niccoli Asabella A, Nicoletti A, Altini C, Notaristefano A, Lastilla G, et al. (2013) 18F-FDG Positron Emission Tomography/Computed Tomography in the Diagnosis and Post-Therapeutic Treatment in a Patient with an Early Stage of Retroperitoneal Fibrosis. Mol Imaging Radionucl Ther 22: 60-62. [Crossref]
- Minford JE, Davies P (1984) The urographic appearances in acute and chronic retroperitoneal fibrosis. Clin Radiol 35: 51-57. [Crossref]
- Benchekroun A, Jira H, Kasmaoui el H, Nouini Y, Iken A, et al. (2002) [Retroperitoneal fibrosis. Report of 6 cases]. Ann Urol (Paris) 36: 171-175. [Crossref]
- Lugosi M, Sacré K, Lidove O, Chauveheid MP, Brihaye B, et al. (2013) [Long-term follow-up of a French cohort of retroperitoneal fibrosis]. Rev Med Interne 34: 591-599. [Crossref]
- Blanc G, Girard N, Alexandre C, Vignon E (2007) Retroperitoneal fibrosis: a rare vascular and immune entity disclosed by chronic lombalgia. Joint Bone Spine 74: 497-499. [Crossref]
- Fibrose rétropéritonéale: une maladie vasculaire ou immunitaire responsable de lombalgie. Retroperitoneal fibrosis: a rare vascular and immune entity disclosed by chronic lombalgia. Revue du Rhumatisme 2007; 74: 902 - 904.
- Tarantino L, Giorgio A, De Stefano G, Esposito F (2000) [Idiopathic retroperitoneal fibrosis diagnosed with color Doppler echography and ultrasonography-guided fine needle biopsy, a case report]. Radiol Med 100: 387-389. [Crossref]
- Ouertani D, Smiti Khanfir M, Klaii R, Braham A, Larbi T, et al. (2009) La fibrose rétropéritonéale. À propos de huit cas. La Revue de médecine interne 30 : S77–S151.
- Mulligan SA, Holley HC, Koehler RE, Koslin DB, Rubin E, et al. (1989) CT and MR imaging in the evaluation of retroperitoneal fibrosis. J Comput Assist Tomogr 13: 277-281. [Crossref]
- Gallais Sérézal I1, Le Jeune S2, Belenfant X3, Bakir R4, Fain O5, et al. (2014) [Idiopathic retroperitoneal fibrosis: a multicentric retrospective study of 30 French cases and follow-up of the renal function]. Rev Med Interne 35: 570-576. [Crossref]
- Triantopoulou C1, Rizos S, Bourli A, Koulentianos E, Dervenis C (2002) Localized unilateral perirenal fibrosis: CT and MRI appearances. Eur Radiol 12: 2743-2746. [Crossref]
- Vivas I, Nicolás AI, Velázquez P, Elduayen B, Fernández-Villa T, et al. (2000) Retroperitoneal fibrosis: typical and atypical manifestations. Br J Radiol 73: 214-222. [Crossref]
- Scheel PJ Jr, Feeley N (2009) Retroperitoneal fibrosis: the clinical, laboratory, and radiographic presentation. Medicine (Baltimore) 88: 202-207. [Crossref]
- Gaertner S, Cordeanu EM, Mirea C, Stephan D (2014) [Retroperitoneal fibrosis]. Presse Med 43: 1021-1023. [Crossref]
- Vaglio A, Versari A, Fraternali A, Ferrozzi F, Salvarani C, et al. (2005) (18)F-fluorodeoxyglucose positron emission tomography in the diagnosis and followup of idiopathic retroperitoneal fibrosis. Arthritis Rheum 53: 122-125. [Crossref]
- Moroni G, Castellani M, Balzani A, Dore R, Bonelli N, et al. (2012) The value of (18) F-FDG PET/CT in the assessment of active idiopathic retroperitoneal fibrosis. Eur J Nucl Med Mol Imaging 39: 1635-1642. [Crossref]
- Desbois AC, Hervier B, Haroche J, Lucidarme O, Wechsler B, Charlotte F, et al. (2010) Fibrose rétropéritonéale « idiopathique » : à propos de 31 cas La Revue de médecine interne 31S (2010) S35-S83
- Drieskens O, Blockmans D, Van den Bruel A, Mortelmans L (2002) Riedel's thyroiditis and retroperitoneal fibrosis in multifocal fibrosclerosis: positron emission tomographic findings. Clin Nucl Med 27: 413-415. [Crossref]
- Estrade V, Traxer O, Sibony M, Haab F (2004) [Retroperitoneal fibrosis]. Ann Urol (Paris) 38: 3-13. [Crossref]
- Kukuk S, Krestschmer A, Bruck H, Roth S, Brandt AS (2015) Retroperitoneal fibrosis: Development of a biomarker profile for diagnosis and therapy monitoring. Urologe A 54: 52-61. [Crossref]
- Ahsan N, Choudhury AA, Berger A (1990) Retroperitoneal fibrosis. Am Fam Physician 41: 1775-1780. [Crossref]
- Klisnick A, Fourcade J, Ruivard M, Baud O, Souweine B, et al. (1999) Combined idiopathic retroperitoneal and mediastinal fibrosis with pericardial involvement. Clin Nephrol 52: 51-55. [Crossref]
- Chauveau D, Fiquet-Kempf B, Mejean A, et al. (1997) Fibrose rétropéritonéale: faits cliniques et physiopathologiques récents. Séminaires d’uronéphrologie. Paris: Flammarion Médecine-Science 226-234.





