In an era of increasing drug resistance, rapid detection and determination of the molecular mechanisms involved in antimicrobial resistance is pivotal for successful treatment of bloodstream infections. The appropriate antimicrobial therapy is crucial to reduce the mortality rate [1] and should be started within 1 hour of sepsis suspicion [2], but empirical therapy is a challenge in those hospitals where resistance mechanisms are an ever-changing landscape [3]. Rapid microorganism identification in positive blood cultures (BCs) has been achieved using a variety of innovative techniques [4-7]. The turnaround time of results has indeed been reduced considerably in the recent years by using the Matrix Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) for pathogen identification in samples taken directly from positive BCs [8,9].
At the Bacteriology Unit of the Pisa University Hospital (Pisa, Italy), Escherichia coli and Klebsiella pneumoniae are the prevalent species isolated from BCs [10]. While in our region, Tuscany, E. coli is still sensitive to most antibiotics, carbapenem resistant strains of K. pneumoniae have developed and spread in the community and in healthcare settings, first as K. pneumoniae strains harbouring the KPC carbapenemase gene (KPC-Kp), and more recently harbouring the NDM gene (NDM-Kp) [11,12]. To guide clinicians in starting the proper therapy, we employed the CarbaR molecular kit (GeneXpert® System, Cepheid, Sunnyvale, CA, USA), which has been developed for the detection of carbapenem resistance mechanisms (blaKPC, blaNDM, blaVIM, blaIMP-1, and blaOXA-48 genes) from rectal swabs or from bacterial colonies. In the present study, we aimed at establishing a rapid method, able to provide detection and differentiation of the most prevalent carbapenemase gene families directly from positive blood cultures containing K. pneumoniae. To this aim, a bacterial pellet was recovered directly from positive BC bottles and partly used for MALDI-TOF MS identification (Bruker Daltonics, Bremen, Germany) according to a previously published protocol [10]; when bacteria were identified as K. pneumoniae, an aliquot of the harvested pellet was used to prepare a 0.5 McFarland suspension. Twenty µl of a 1:10 dilution in distilled water were inoculated in the Xpert® Carba-R lysis buffer and vigorously vortexed for 1 minute. Next, an aliquot (1.7 ml) was transferred into the Xpert® Carba-R cartridge for detection and differentiation. Blood samples from patients admitted to the Pisa University Hospital in the period June 2016 - May 2019 were inoculated into blood culture bottles Plus Aerobic/F and Plus Anaerobic/F, or Peds Plus F (Becton Dickinson &Co, BD, Milan, Italy). In total, 101.290 BCs were collected at the Microbiology Unit and transferred to the Bactec FX Instrument (Becton Dickinson, Franklin Lakes, NJ, USA), 10.405 of which became positive. Among the positive BC bottles, 965 were for K. pneumoniae. For each patient, only the first positive BC bottle was processed by the method herein described: in total, 406 K. pneumoniae were directly identified by MALDI-TOF MS in about 30', while the molecular detection of genes involved in carbapenem resistance took 48'. Carbapenem resistance mechanisms were found in 166 (40.9%) BCs. All strains were subsequently analysed phenotypically reaching 100% agreement with the molecular method. As shown in figure 1, the proportion of KPC-Kp producer was 45% in 2016, 43% in 2017, 31% in 2018, and 25% in 2019. Both in 2016 and in 2017, two strains of K. pneumoniae harbouring carbapenemase VIM gene were detected; K. pneumoniae harbouring carbapenemase OXA-48 gene was detected only once in 2016. NDM-Kp was not detected in 2016 and 2017, whereas it was detected with a percentage of 3% in 2018, and 20% in 2019 (January-May) [12]. K. pneumoniae harbouring carbapenemase IMP-1 gene has not yet been detected.
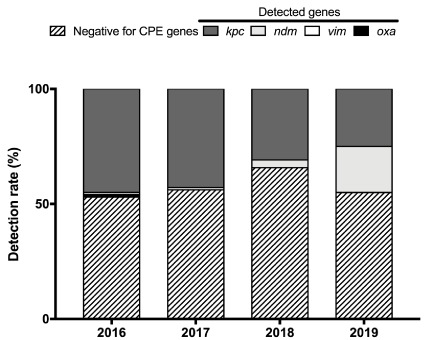
Figure 1. Blood culture trend for Carbapenamase-producing Enterobacterales (CPE)
In Northern Tuscany, we witnessed a progressive evolvement over time of resistance mechanisms to carbapenems, and an overpowering appearance of NDM-Kp. In July 2019, our regional health directorate issued a strict protocol for the surveillance and control of infections, in an attempt to stem the spread of the epidemic of resistant strains. In this context, a rapid method for detecting molecular mechanisms of resistance to carbapenems, directly from positive BCs, may be crucial for infection management. Indeed, patients with positive results could be promptly isolated, and differently isolated, depending on the detected mechanism of resistance. This method requires about 78’ for rapid MALDI-TOF MS identification of K. pneumoniae and detection and differentiation of carbapenem resistance mechanisms. In conclusion, we believe that this method allows both to design effective infection control measures and to promptly administer the correct antimicrobial therapy, as it is known that the use of new drugs, eg. ceftazidime/avibactam is not effective against NDM-Kp.
carbapenem resistance, blood cultures, New Delhi metallo-β-lactamase, NDM, MALDI TOF MS
- Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, et al. (2006) Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34: 1589-1596. [Crossref]
- Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, et al. (2017) Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med 43: 304- 377. [Crossref]
- Peralta G, Sanchez MB, Garrido JC, De Benito I, Cano ME, et al. (2007) Impact of antibiotic resistance and of adequate empirical antibiotic treatment in the prognosis of patients with Escherichia coli bacteraemia. J Antimicrob Chemother 60: 855-863. [Crossref]
- Salimnia H, Fairfax MR, Lephart PR, Schreckenberger P, DesJarlais SM, et al. (2016) Evaluation of the filmarray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol 54: 687-698. [Crossref]
- Arroyo MA, Denys GA (2017) Parallel evaluation of the maldi Sepsityper and verigene BC-GN assays for rapid identification of gram-negative bacilli from positive blood cultures. J Clin Microbiol 55: 2708-2718. [Crossref]
- Kothari A, Morgan M, Haake DA (2014) Emerging technologies for rapid identification of bloodstream pathogens. Clin Infect Dis 59: 272-278. [Crossref]
- Kim JS, Kang GE, Kim HS, Kim HS, Song W, et al. (2016) Evaluation of Verigene Blood Culture Test Systems for Rapid Identification of Positive Blood Cultures. Biomed Res Int 1081536. [Crossref]
- Barnini S, Ghelardi E, Brucculeri V, Morici P, Lupetti A (2015) Rapid and reliable identification of Gram-negative bacteria and Gram-positive cocci by deposition of bacteria harvested from blood cultures onto the MALDI-TOF plate. BMC Microbiology 15: 124. [Crossref]
- Mauri C, Principe L, Bracco S, Meroni E, Corbo N, et al. (2017) Identification by mass spectrometry and automated susceptibility testing from positive bottles: a simple, rapid, and standardized approach to reduce the turnaround time in the management of blood cultures. BMC Infect Dis 17: 749. [Crossref]
- Giordano C, Piccoli E, Brucculeri V, Barnini S (2018) A Prospective Evaluation of Two Rapid Phenotypical Antimicrobial Susceptibility Technologies for the Diagnostic Stewardship of Sepsis. Biomed Res Int 10: 6976923. [Crossref]
- ARS Toscana, Agenzia Regionale di Sanità. L'antibiotico resistenza e l'uso di antibiotici in Toscana. www.ars.toscana.it. 2019. ISSN 1970-3252.
- ECDC. 4 June 2019. Rapid Risk Assessment: Regional outbreak of New Delhi metallo-betalactamase-producing carbapenem-resistant Enterobacteriaceae, Italy, 2018–2019. https://ecdc.europa.eu/en/publications-data/RRA-new-delhi-metallo-beta-lactamase-producing-CRE.

