Background: To reduce over diagnosis and overtreatment of prostate cancer, a noninvasive and easy to administer test is needed to assess the risk of clinically significant disease. Such an assay must also be able to help accurately inform whether a prostate biopsy is warranted.
Objective: To determine the performance of a novel, serum-based multiplexed autoantibody assay vs. PSA for discriminating high-grade prostate cancer on biopsy (Gleason Score 7 or greater).
Methods: Using retrospective serum samples, we compared the autoantibody assay results with biopsy outcomes in 760 patients (an autoantibody-binding peptide discovery study of 268 samples, a training set of 196 samples and a validation cohort comprised of 296 samples) with or at risk of prostate cancer from two academic sites and one community clinical facility in the United States. Eligible participants included men aged 40 years or older, undergoing a prostate biopsy due to suspicious digital rectal examination finding and/or elevated PSA level.
Results: Among the 196 men in the training set (average age 61.9, average PSA 7.4 ng/mL), the autoantibody assay plus age showed better discrimination (Area Under the Curve (AUC) 0.74, 95% CI (0.68-0.80)) when compared to PSA alone (AUC 0.56, 95% CI (0.54-0.68)). Based on an assumed clinical decision for patients falling below the test cut point, 31.6% of unnecessary biopsies could have been avoided.
Among the 296 men in the validation study (average age 63.3 years, range 40-90; average PSA 6.0 ng/mL), the autoantibody assay showed better discrimination (AUC 0.69, 95% CI 0.63-0.75) than PSA (AUC 0.55, 95% CI 0.48-0.62) (P =0.007) for separating Gleason Score 7 or greater from Gleason Score 6 and patients negative on biopsy. For detected Gleason Score 7 or higher test with 95% sensitivity, 20.1% of unnecessary biopsies could have been avoided, missing only 4.9% of patients with Gleason Score 7 disease. Of these 6 patients whose cancers were missed, a sub-analysis revealed five were Gleason 7 (3+4) and only one was Gleason 7 (4+3).
Conclusion: This study evaluated a novel cancer-specific biomarker assay based on autoantibody signatures that could be used as a noninvasive risk assessment aid for high-grade prostate cancer. When results of this assay are combined with traditional clinical risk factors, risk stratification for high-grade prostate cancer and biopsy decision making are improved compared to current methods in clinical practice. We hypothesize patients with Gleason Score 6 will exhibit the greatest benefits from employing the assay as they may likely avoid both unnecessary prostate biopsy and subsequent overtreatment. We further hypothesize the assay will thus significantly reduce costs to the healthcare system while further improving patient’s quality of care. Providers and their patients suspected of having prostate cancer may consider using this novel assay prior to proceeding with prostate biopsy.
prostate cancer, cancer-specific biomarker assay, prostate biopsy
Prostate cancer is the most common non-skin related cancer among men in the United States, with an estimated 161,360 new cases of prostate cancer and 26,730 deaths from prostate cancer in 2017 [1].
While many prostate cancer cases will not progress, or will progress slowly and remain asymptomatic during a lifetime, select cases are aggressive and warrant early detection and treatment [2].
Deaths from prostate cancer have declined by approximately 40% since the advent of prostate-specific antigen (PSA) screening in the late 1980s [3]. However, the PSA test lacks specificity for high-grade disease, leading to a high rate of false-positive results and unnecessary, repeat biopsies, which pose the risk for bleeding, infection, and pain [2,4]. In addition, harms of over diagnosis and treatment, stemming from PSA testing include, infection, blood loss requiring transfusion, pneumonia, erectile dysfunction, and incontinence [5]. Furthermore, patient anxiety may result from false-positive results of PSA testing as well as having to undergo prostate biopsy [6].
The uncertain benefit of PSA-based screening, combined with the complications associated with standard of care with treatment, led the U.S. Preventive Services Task Force (USPSTF) in 2012 to recommend against regular screening [2]. While this recommendation reduced the absolute number of prostate needle biopsies performed by 31%, it is also linked to a reduction in the detection of intermediate risk tumors, which are potentially curable [7]. In addition, research shows that the incidence of metastatic prostate cancer has risen significantly – by 72% -- between 2004 and 2013, with a 92% increase among men aged 55 to 69 years old [8].
Recently, the USPSTF has amended its stance in a draft statement [9], determining that the decision about whether to be screened should be an individual one. The new recommendation –which changed the grade for PSA screening from D to C – states the potential benefits and harms of PSA-based screening are closely balanced in men aged 55 to 69 years. As a result, the USPSTF recommends clinicians evaluate a person’s risk because of family history and race as well as competing causes of morbidity and mortality. Talking to these men about the potential benefits and harms of screening is encouraged; if PSA is measured and found to be increased, then substantial conversation needs to ensue to help the patient decide on the next step. This approach aligns more closely with the current recommendations of the American Urological Association [10].
Controversy surrounding the USPSTF recommendation and the limitations of the PSA test have spurred research on novel serum and tissue biomarkers to identify patients at risk for intermediate- or high-risk prostate cancer. Several other serum, urine, or biopsy tissue-derived biomarkers are available to aid in prostate cancer diagnosis, but unlike the serum-based novel multiplexed autoantibody assay described herein, many are based in some way on measurement of PSA. Efforts toward the development of screening tests for prostate cancer have generally depended on single biomarker molecules, primarily PSA, as well as PCA3 [11]. Current technologies have been disappointing and have not resulted in diagnostic tests sufficiently reliable or convenient to apply to clinical practice for early detection of aggressive prostate cancer. Consequently, there is a need for new biomarkers that can identify prostate cancer at any state of prostate cancer progression while limiting the number of false positives [12]. This is especially true in the intermediate range of PSA detection (4-10 ng/mL) [12].
The discovery that patients with cancer produce detectable autoantibodies against antigens in their tumors suggests that these biomarkers could have diagnostic and prognostic value [13]. Building upon these findings, researchers identified a panel of eight peptides that are recognized by autoantibodies generated by the immune system in response to the presence of prostate cancer and developed an algorithm that can be used to indicate a relatively high or low risk of prostate cancer, particularly for patients with intermediate (4.0 to 10 ng/mL) PSA levels [12].
This non-PSA blood test – known as Apifiny®-- has demonstrated efficacy in identifying men at high risk of prostate cancer in a variety of clinical studies [12]. Apifiny® has the potential to aid clinicians in determining the most appropriate candidates for an initial or repeat biopsy and is intended for use in patients with prostate cancer risk factors to help provide additional insight to support a more informed clinical decision about performing a prostate biopsy.
Most recently, researchers have examined the performance of the 8 Apifiny® phage peptides when used in combination with an extended set of up to 13 new autoantibody-binding peptides in detecting high- grade prostate cancer on biopsy (i.e., Gleason Score ≥ 7) [14]. The aim of this study was to validate the performance of this expanded serum-based multiplexed autoantibody assay in the risk assessment of patients with high-grade prostate cancer compared to PSA.
Validation study population
Retrospective serum samples and annotated clinical information for 296 biopsy confirmed patients were sourced from two academic sites and one community facility in the United States. Eligible participants included men aged 40 years or older, undergoing a prostate biopsy due to suspicious digital rectal examination finding and/or elevated PSA level. Only men with previously diagnosed prostate cancer were excluded from the study.
The men had an average age of 63.3 and an average PSA of 6.0 ng/mL. Twenty-nine percent of men had a PSA <4.0 ng/mL, 60% had a PSA between 4 to 10 ng/mL, and only 10% of men had a PSA greater than 10 ng/mL. The majority (61%) of the men were Caucasian, 6% were African American and in 32% of samples, no race was specified; they were therefore classified as ‘other’. Twenty-seven percent of men had Gleason 6 cancer, 29% had Gleason 7 (3+4) cancer, 7% had Gleason 7 (4+3) cancer, and 20% of men had Gleason 7 cancer with no breakout. Finally, 17% of men had Gleason 8 or greater. Almost all men (98%) in the biopsy positive group had only one biopsy. Put another way, only 2% or a total of three (3) men, underwent repeat biopsy. In the control group, 70% of men had one biopsy, 12% had two biopsies, 8% had three biopsies, and 10% of men had four or more biopsies (Tables 1-5).
Table 1. Age Ranges in the Training Set and the Validation Study
AGE Ranges: Training Set |
AGE Ranges: Validation Study |
Positive Samples |
Control Samples |
TOTAL |
Positive Samples |
Control Samples |
TOTAL |
40-55 |
17 |
19% |
40-55 |
24 |
23% |
41 |
21% |
40-55 |
22 |
15% |
40-55 |
21 |
14% |
43 |
15% |
> 55 to 65 |
43 |
48% |
> 55 to 65 |
52 |
49% |
95 |
48% |
> 55 to 65 |
72 |
50% |
> 55 to 65 |
73 |
48% |
145 |
49% |
> 65 to 75 |
20 |
22% |
> 65 to 75 |
25 |
24% |
45 |
23% |
> 65 to 75 |
35 |
24% |
> 65 to 75 |
49 |
32% |
84 |
28% |
> 75 |
10 |
11% |
> 75 |
5 |
5% |
15 |
8% |
> 75 |
14 |
10% |
> 75 |
10 |
7% |
24 |
8% |
TOTAL |
90 |
|
TOTAL |
106 |
|
196 |
|
TOTAL |
143 |
|
TOTAL |
153 |
|
296 |
|
AVG Age |
63 |
|
AVG Age |
61 |
|
|
|
AVG Age |
63 |
|
AVG Age |
63 |
|
|
|
Table 2. PSA Levels in the Training Set and the Validation Study
PSA Levels: Training Set |
PSA Levels: Validation Study |
Positive Samples |
Control Samples |
TOTAL |
Positive Samples |
Control Samples |
TOTAL |
less than 4 |
18 |
20% |
less than 4 |
36 |
34% |
54 |
28% |
less than 4 |
33 |
23% |
less than 4 |
53 |
35% |
86 |
29% |
4 to 10 |
57 |
63% |
4 to 10 |
57 |
54% |
114 |
58% |
4 to 10 |
92 |
64% |
4 to 10 |
87 |
57% |
179 |
60% |
> 10 |
15 |
17% |
> 10 |
13 |
12% |
28 |
14% |
> 10 |
18 |
13% |
> 10 |
13 |
8% |
31 |
10% |
TOTAL |
90 |
|
TOTAL |
106 |
|
196 |
|
TOTAL |
143 |
|
TOTAL |
153 |
|
296 |
|
AVG PSA |
8.0 |
|
AVG PSA |
6.0 |
|
|
|
AVG PSA |
6.7 |
|
AVG PSA |
5.3 |
|
|
|
Table 3. Race Breakout in the Training Set and the Validation Study
|
Race Breakout: Training Set |
Race Breakout: Validation Study |
|
Positive Samples |
Control Samples |
Total |
Positive Samples |
Control Samples |
Total |
African American |
8 |
9% |
1 |
1% |
9 |
5% |
15 |
10% |
4 |
3% |
19 |
6% |
Caucasian |
76 |
84% |
45 |
42% |
121 |
62% |
118 |
83% |
63 |
41% |
181 |
61% |
Other |
6 |
7% |
60 |
57% |
66 |
34% |
10 |
7% |
86 |
56% |
96 |
32% |
Total |
90 |
|
106 |
|
196 |
|
143 |
|
153 |
|
296 |
|
Table 4. Gleason Scores in the Training Set and the Validation Study
|
Gleason Scores: Training Set |
Gleason Scores: Validation Study |
Six |
26 |
29% |
38 |
27% |
Seven (3+4) |
27 |
30% |
42 |
29% |
Seven (4+3) |
6 |
7% |
10 |
7% |
Seven (w/out breakdown) |
13 |
14% |
29 |
20% |
Eight |
7 |
8% |
7 |
5% |
Nine + |
11 |
12% |
17 |
12% |
TOTAL |
90 |
100% |
143 |
100% |
Table 5. Number of Biopsies in the Training Set and the Validation Study
Number of Biopsies: Training Set |
Number of Biopsies: Validation Study |
Positive Samples |
Control Samples |
Positive Samples |
Control Samples |
# of Biopsies |
# of Samples |
% |
# of Biopsies |
# of Samples |
% |
# of Biopsies |
# of Samples |
% |
# of Biopsies |
# of Samples |
% |
1 |
89 |
99% |
1 |
84 |
79% |
1 |
140 |
98% |
1 |
107 |
70% |
2 |
1 |
1% |
2 |
12 |
11% |
2 |
2 |
1% |
2 |
19 |
12% |
3 |
0 |
0% |
3 |
9 |
8% |
3 |
0 |
0% |
3 |
12 |
8% |
4 |
0 |
0% |
4 |
1 |
1% |
4 |
1 |
1% |
4 |
15 |
10% |
Total |
90 |
|
Total |
106 |
|
Total |
143 |
|
Total |
153 |
|
Sample collection and processing
Retrospective samples for these cohorts were obtained from three different sources (the University of Michigan, Johns Hopkins University, and the Michigan Institute of Urology). All samples in the autoantibody-binding peptides discovery study, the training set and the validation study were prostate cancer positive and biopsy negative controls and all cohorts were age and PSA matched.
Samples were shipped to a central laboratory and processed or stored upon receipt.
No patients were compensated for participating. Test results were not provided to the clinical sites for patient care, and the laboratory technicians who performed the biomarker tests were blinded for patient characteristics. The autoantibody-binding peptides discovery study, the training set study and the validation study were all performed in accordance with the Standards for Reporting of Diagnostic Accuracy criteria [15].
Laboratory-developed test: Novel serum based multiplexed autoantibody assay development
State-of-the-art genomic and proteomic information was utilized to develop and refine an assay that can quantify the presence and aggressiveness of prostate cancer (Figure 1). A total of 268 samples in the autoantibody-binding peptide discovery and assessment study were collected from prostate cancer positive and biopsy negative controls. Twenty-one phage-peptides were analyzed to determine the specific biological markers known to be associated with an immune system response to aggressive prostate cancer [14].
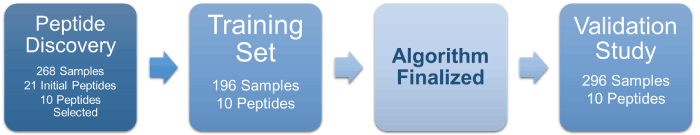
Figure 1. Assay Development and Validation Process.
The phage proteins were developed by a technique in which bacteriophages are engineered to fuse a foreign peptide or protein with their capsid proteins and hence expose or display it on their external surface [13]. The immobilized phage may then be used as a screen to see which ligands bind to the expressed fusion protein displayed on the phage surface. The bio-panning process is an iterative process of affinity purification used in the identification and isolation of phages carrying peptides with high affinity and specificity to a given target molecule. Briefly, a library of phage-displayed peptides is incubated with a plate (or bead substrate) coated with the target. Unbound phage particles are washed away, and the bound phages eluted. This process is then repeated several times. This process was used to develop the library of autoantibody-binding peptides used in the biomarker discovery phase of product development
Our aim was to then test and validate the performance of this expanded serum-based multiplexed autoantibody assay in the risk assessment of patients with high-grade prostate cancer versus PSA. Using a training set of 196 subjects, a unique algorithm was developed to discern between men with low- and high-grade cancers independent of PSA levels. The final algorithm incorporated the ten autoantibody- binding peptides prioritized for aggressive cancer detection (Table 6).
Table 6. Aggressive Cancer Autoantibody-Binding Peptides and their Functions
Autoantibody-Binding Peptides |
Function |
ADP-Ribosylation Factor 6 (ARF 6) |
regulates actin cytoskeleton remodeling vesicle shedding by tumor cells |
NK3 homeobox 1 (NKX3-1) |
regulates androgen response genes (BMI1) |
5’-UTR-BMI1 |
androgen response gene |
3’- UTR- Ropporin |
responsible for ciliary movement in spermatozoa through dynein regulation |
Aurora Kinase Interacting Protein 1 (AURKAIP-1) |
regulates androgen response genes (TWIST1) |
Casein Kinase 2, alpha prime polypeptide (CSNK2A2) |
regulates cell cycle and cellular division |
hnRNPA1 |
up regulated in PCa |
KDELR1 |
SRC pathway regulator |
SMAD3 |
tumorgenesis |
BRD2 |
tanscription regulator |
FKBP4 |
chaperon protein |
The assay and the classifier algorithm using the 10 specific autoantibody-binding peptides optimizes the negative predictive value and minimizes false negative results for Gleason 7+ disease. Finally, a validation analysis using independent samples from 296 subjects assessed the performance of the diagnostic algorithm.
Statistical analysis
The primary objective was to validate the accuracy of the serum-based multiplexed autoantibody assay to predict high-grade prostate cancer for men aged 40 years or older.
These studies fully complied with the recommendations of the 2012 report from the Institute of Medicine Committee on Omics-Based Predictive Tests [16]. Results of the studies were analyzed by independent biostatisticians.
Because the immune system continuously changes as cancer risk fluctuates, the performance of the peptides was assessed and evaluated as area under the curve (AUC) of the receiver operating characteristic (ROC) curve. The 95% confidence intervals (CIs) and comparisons of AUCs were determined using DeLong’s method as implemented in the R package pROC. The combination and predictive value of the multiplexed autoantibody assay was modeled by logistic regression analysis. The assay aimed to identify high-grade (Gleason Score ≥ 7) prostate cancer. The control group included men who either did not have cancer or men who had low-grade cancer (Gleason Score ≤ 6), wherein active surveillance (i.e. no treatment) is the preferred management strategy [17]. The logistic regression model was then applied to estimate the probability of high-grade prostate cancer on biopsy.
Using T7 phage with epitope inserts, autoantibodies against peptides/inserts derived from prostate cancer were identified. At the completion of the biomarker discovery study, samples were randomly divided into a training set (N = 196) and a validation set (N = 296). These two sets were checked for comparability with respect to age, race, and the date each sample was run. Raw biomarker values were normalized against an internal standard control sample. Logistic regression models were used to model the probability of a sample being high grade cancer as a function of the potential biomarkers. ROC curves based on fitted multivariate logistic regression models, were also generated to show the relation between sensitivity and specificity for the range of possible cut points.
To determine the optimal number of peptides to include in the final model, we used the following process. For each number of peptides N= 1 to 21 and each normalization method (a total of two including unnormalized) the model with the highest discrimination (based on AUC) with N peptides was calculated. No interactions were considered. Although the cross-validated estimates of AUC were similar for a model with twelve peptides, the numerically highest cross-validated estimate of AUC was obtained with ten peptides using the T7 ratio normalization method. To normalize the peptides with T7, for each subject we divided each peptide by that subject's T7 value. Thus, the final model selected for validation was obtained as the best discriminating model (on the training data) containing ten peptides using the T7 ratio normalization.
An estimate of predicted risk was defined as the linear combination of the ten selected peptides each multiplied by their parameter coefficient from the fitted logistic regression model. This risk was calculated for patients in the validation set, and a table of model performance statistics was generated using each risk estimate in the validation data set as a decision threshold for the prediction of high- grade cancer. Specifically, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated at all present cut points. In addition, PPV and NPV were calculated using test sensitivity and specificity at the US average incidence of prostate cancer defined prevalence of 18%. All analysis was performed using R 3.32 (R Core Team, Vienna, Austria).
Selection of the most informative aggressive cancer autoantibody-binding peptides
The performance of individual peptides to predict high-grade prostate cancer was compared. In addition to the AUC, specificity, negative predictive value (NPV), and positive predictive value (PPV) were determined at a fixed sensitivity of approximately 90%. A single cut point with a negative predictive value (NPV) of greater than 95% was selected. Figure 2 shows for Waterfall Plot of the Autoantibody Assay Score across the Validation cohort. Negative predictive value was optimized to minimize the risk of missing an aggressive cancer. The statistical analysis demonstrated the peptides have a degree of relatedness. Indeed, the peptides do not function as stand-alone autoantibody- quantifiers, but instead they rely and depend on each other to perform. The combination of these 10 biomarkers had the best performance (AUC 0.69, 95% CI 0.63-0.75) compared to PSA alone (AUC 0.55, 95% CI 0.48-0.62) (P =0.007) (Figure 3).
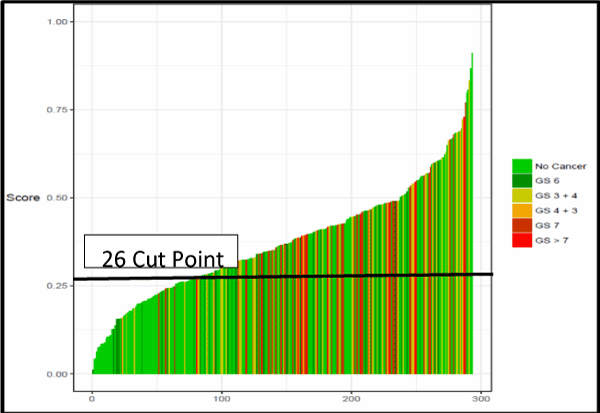
Figure 2. Waterfall Plot of the Autoantibody Assay Score Across the Validation Cohort
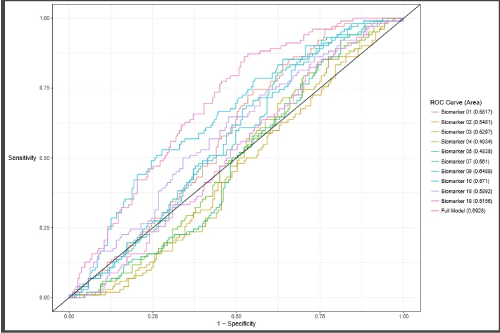
Figure 3. Receiver Operating Curve from the Validation Study.
Clinical validation
The performance characteristics of the assay were evaluated relative to PSA in the independent validation study. Among the 296 men (median age 63.3 years; median PSA 6.0 ng/mL), the expanded autoantibody assay showed significantly better discrimination between patients with Gleason Score 7 or greater from Gleason Score 6 and patients negative on biopsy (AUC 0.69, 95% CI 0.63-0.75) compared to PSA alone (AUC 0.55, 95% CI 0.48-0.62) (P =0.007). A test developed with this expanded set of phage- peptide clones detected Gleason Score 7 or higher with 95.0% sensitivity and 28% specificity. For biopsies with a Gleason Score of ≥ 7, approximately 20.1% of unnecessary biopsies could potentially have been avoided, missing only 4.9% of patients with high-risk cancer, Gleason Score 7 disease. A sub analysis of the six (6) aggressive cancers missed found five to be Gleason 7 (3 + 4); only one (1) was Gleason 7 (4+3). A rigorous statistical analysis resulted in a single cut point of 26; at this point, negative predictive value was optimized at 92%. The positive predictive value was 42%. At this cut point, the percentage of false negatives was 1.7%. The validation study population prevalence rate was 35.5% for aggressive disease. Thus, this expanded multiplexed autoantibody assay provides significant clinical benefit when compared with standard clinical tools (Table 7).
Efforts toward the development of screening tests for prostate cancer have generally depended on single biomarker molecules, primarily PSA, as well as PCA3 [11]. Current technologies have been disappointing and have not resulted in diagnostic tests sufficiently reliable or convenient to apply to clinical practice for early detection of aggressive PCa. Consequently, there is a need for new biomarkers that can identify prostate cancer at any state of progression while limiting the number of false positives [12]. This is especially true in the intermediate range of PSA detection (4-10 ng/mL) [12].
In the landmark Prostate Cancer Prevention Trial (PCPT) [18], Thompson et al reported the diagnosis of prostate cancer in 15.2% of men with a PSA level of ≤ 4 ng/mL. Furthermore 14.9% of those men were found to harbor high-grade disease [18].
As articulated by Schipper and colleagues [12], multiple complex molecular events characterize prostate cancer initiation, unregulated growth, invasion, and metastasis [12]. Distinct sets of genes and proteins dictate progression from precursor lesion to localized disease and finally to metastatic disease [12].
Biomarkers that detect prostate cancer in any of these states of progression would be ideal as we now more fully understand there is an immune response to cancer in humans which has been demonstrated by the identification of autoantibodies against a number of intracellular antigens in patients with various tumor types [12]. This phenomenon is known as the humoral response and the detection of such autoantibodies has been shown to have great diagnostic and prognostic value in the detection of cancer and the ability to predict the course of the disease [12]. In addition, it has been shown that many antigens from tumor cells that elicit a response are not just products of mutated genes. These proteins are often differentiation antigens or other proteins over-expressed in cancer [12].
Using iterative biopanning and phage-protein microarrays, Wang et al. [13] developed an assay whereby multiple autoantibody biomarkers can be used to screen for prostate cancer. An algorithm was developed to discern healthy from diseased individual’s independent of PSA [13]. Clinical studies [12] show by relying on multiple immunogenetic prostate-cancer peptides, APIFINY’s novel approach appears to be a significant improvement over a single biomarker such as PSA, as it has been shown to improve diagnosis of prostate using a biologic measure independent of PSA. Such markers will more accurately identify patients who are most likely to benefit from referral to a urologist for further evaluation, biopsy and, potentially, treatment for early prostate cancer while reducing inaccurate readings, unnecessary invasive testing in healthy men, and associated morbidity and healthcare costs.
This study assessed the performance of a novel serum-based multiplexed autoantibody assay using 10 phage-peptide clones compared to PSA alone for detecting high-grade prostate cancer on biopsy (Gleason Score 7 or greater). The algorithm derived from this model was the best performing assay to detect high-grade prostate cancer on prostate biopsy and was successfully validated in an independent cohort of 296 samples. The assay significantly outperformed PSA alone. Moreover, the algorithm used to generate the performance does not require PSA, its derivatives or any other factors in the assay to be effective. As well, age has no effect on the assay biomarker performance. Thus, when a patient’s score is below the cut point, there is a decreased likelihood of high-grade cancer. Put another way, scores below 26 indicate a 92% predicted probability of not having aggressive prostate cancer. Consequently, when combined with currently acceptable methods of risk assessment, the results of this test enable objective clinical risk assessment and patient management.
The current study has several limitations, including the need to more fully assess the test in other races, and how other conditions (such as obesity and its pro-inflammatory state, or steroid use) may affect the assay’s performance. As well, the outcome of this study was ≥ G7 on biopsy; it is known some G7 disease is indolent, and some, high-volume G6 is clinically meaningful. Finally, this study’s outcome was biopsy; it is well documented that approximately 25% of biopsies will return a false-negative result [19].
Ideally, long-term follow-up including prostate cancer death are needed to verify this as a marker of aggressive prostate cancer.
This study evaluated a novel cancer-specific biomarker assay based on autoantibody signatures that could be used as an easy to administer, noninvasive risk assessment aid for high-grade, clinically significant prostate cancer. When results of this assay are combined with traditional clinical risk factors, risk stratification for high-grade prostate cancer and biopsy decision making are improved. We hypothesize patients with Gleason Score 6 will exhibit the greatest benefits from employing the assay as they will likely avoid both unnecessary prostate biopsy and subsequent overtreatment. Thus, the assay described herein may significantly reduce costs to the healthcare system while improving patient’s quality of care. Further study is encouraged to test this theory. In the meantime, however, based on the results of this study, providers and their patients suspected of having prostate cancer may consider using this novel assay prior to proceeding with prostate biopsy.
We are indebted to Arul Chinnaiyan, M.D., Ph.D. for his continued encouragement of this work, to Dr. Ashley Ross and Dr. Robert Reinhardt for their guidance, and to Jim Corrigan and the entire Armune Bioscience staff for their assistance in the laboratory.
MagArray Inc provided all financial support for the work reported in this article.
- American Cancer Society (2017) Key Statistics for Prostate Cancer. https://www.cancer.org/cancer/prostate-cancer/about/key-statistics.html. Accessed March 23, 2017.
- Moyer VA, U.S. Preventive Services Task Force (2012) Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 157: 120-134. [Crossref]
- Hugosson J, Carlsson S, Aus G, Bergdahl S, Khatami A, et al. (2010) Mortality results from the Goteborg randomised population-based prostate-cancer screening trial. Lancet Oncol 11: 725-732. [Crossref]
- Rosario DJ, Lane JA, Metcalfe C, Donovan JL, Doble A, et al. (2012) Short term outcomes of prostate biopsy in men tested for cancer by prostate specific antigen: prospective evaluation within ProtecT study. BMJ 344: d7894. [Crossref]
- Ilic D, Neuberger MM, Djulbegovic M, Dahm P (2013) Screening for prostate cancer. Cochrane Database Syst Rev: CD004720. [Crossref]
- Carlsson S, Aus G, Wessman C, Hugosson J (2007) Anxiety associated with prostate cancer screening with special reference to men with a positive screening test (elevated PSA) - Results from a prospective, population-based, randomised study. Eur J Cancer 43: 2109-2116. [Crossref]
- Banerji JS, Wolff EM, Massman JD 3rd, Odem-Davis K, Porter CR, et al. () Prostate Needle Biopsy Outcomes in the Era of the U.S. Preventive Services Task Force Recommendation against Prostate Specific Antigen Based Screening. J Urol 195: 66-73. [Crossref]
- Weiner AB, Matulewicz RS, Eggener SE, Schaeffer EM (2016) Increasing incidence of metastatic prostate cancer in the United States (2004-2013). Prostate Cancer Prostatic Dis 19: 395-397. [Crossref]
- https://www.urotoday.com/recent-abstracts/urologic-oncology/prostate-cancer/95197-u-s-preventive-services-task-force-prostate-cancer-screening-draft-recommendations.html
- Carter HB, Albertsen PC, Barry MJ, Etzioni R, Freedland SJ, et al. (2013) Early detection of prostate cancer: AUA Guideline. J Urol 190: 419-426. [Crossref]
- Jacobsen SJ, Katusic SK, Bergtralh EJ, Oesterling JE, Ohrt D, et al. (1995) Incidence of prostate cancer diagnosis in the eras before and after serum prostate-specific antigen testing. JAMA 274: 1445-1449. [Crossref]
- Schipper M, Wang G, Giles N, Ohrnberger J (2015) Novel prostate cancer biomarkers derived from autoantibody signatures. Transl Oncol 8: 106-111. [Crossref]
- Wang X, Yu J, Sreekumar A, Varambally S, Shen R, et al. (2005) Autoantibody signatures in prostate cancer. N Engl J Med 353: 1224-1235. [Crossref]
- Freedland S, Singh S, Kapphahn K, Howard L, Ohmberger J, et al. (2017) A novel serum based multiplexed 21 autoantibody assay to predict high-grade prostate cancer at initial biopsy. J Urol 197: e340.
- Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, et al. (2003) The STARD statement for reporting studies of diagnostic accuracy: explanation and elaboration. Ann Intern Med 138: W1-12. [Crossref]
- Micheel CM, Nass SJ, Omenn GS, et al. (2012) Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington (DC): National Academies Press (US). [Crossref]
- https://www.auanet.org/guidelines/clinically-localized-prostate-cancer-new-(aua/astro/suo-guideline-2017)
- Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, et al. (2004) Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med 350: 2239-2246. [Crossref]
- Gershman B, Van Houten HK, Herrin J, Moreira DM, Kim SP, et al. (2017) Impact of Prostate-specific Antigen (PSA) Screening Trials and Revised PSA Screening Guidelines on Rates of Prostate Biopsy and Postbiopsy Complications. Eur Urol 71: 55-65. [Crossref]



