The search for bioactive compounds against cancer cell lines produced by endophytes led to the isolation of an actinomycete identified by 16S rDNA as Streptomyces scabrisporus. The organic extracts of this microorganism were evaluated in a bioassay-guided study for cytotoxic activity against breast adenocarcinoma MCF7 and human cervical carcinoma HeLa cell lines. The major and also more active compound was isolated and completely characterized as steffimycin-B (StefB). This compound showed cytotoxic activity against HeLa cells and caused morphological changes in MCF-7 cells with an IC50 value of 2.56 μM. StefB also affected other breast cancer cell lines, including the triple negatives HCC1806 and DU4475 (IC50 values of 18.2 and 4.82 μM, respectively) and exhibited low activity against the non-cancerous line of keratinocytes HaCaT (IC50 156 μM). Besides, StefB decreased expression of the proliferating cell nuclear antigen (PCNA), induced morphological changes and apoptosis on MCF7 cells, which increased with the exposure time, supporting this compound as a promissory drug for breast cancer treatment.
Amphipterygium adstringens, Streptomyces scabrisporus, endophytes, steffimycin B, cytotoxicity
Screening and isolation of soil Actinomycetes with potential pharmacological properties has been explored for more tan 50 years. Owing to the movement of the pharmaceutical industry away from natural products, in recent years the number of drug approvals has drastically dropped down [1]. However, microbial products with pharmaceutical applications are still big business and the search for new drugs cannot be stopped. Therefore, new screening approaches including new metagenomic and genome-mining techniques and the exploration of non-conventional sources of potentially producer microorganisms, are still being studied [2].
Endophytes are microorganisms that inhabit plants without causing visible disease symptoms [3]. Along the past few years they have become an important source of novel molecules with interesting biological characteristics [4].
Discovery of anticancer compounds in endophytes started with the isolation of taxol from Taxomyces andreanae, a molecule widely used in chemotherapy [5]. Towards 2010, 57 fungal and 13 bacterial endophytes producing taxol or some of its precursors were reported [6], reaching over 80 fungi by 2014 [7,8]. A similar story has been observed for other commercially important anticancer compounds. Thus, 11 fungal endophytes able to produce camptothecin and 14 yielding podophyllotoxin, were isolated from different host-plants [9-11].
Diversification of novel compounds produced by endophytes increases every day. Thus, 100 anticancer molecules were isolated between 1990 and 2010, belonging to 19 different chemical classes and showing activity against 45 different cell lines [11]. Additional compounds recently isolated from endophytes exhibited antibiotic [12], antifungal [13], antiparasitic [14] and antiviral activities [15].
Actinomycetes have the potential to produce a huge number of compounds that differ greatly between genera. Some of them contain more than 60 clusters for secondary metabolites, mostly unknown [16]. Screening and isolation of endophytic actinomycetes is important, due to the interaction and evolution developed between host plants and endophytes. The use of medicinal plants as source of actinomycetes has gained interest due to the biotechnological potential in the production of different and novel compounds [17].
The plant selected for this study was Amphipterygium adstringens, traditionally known as "Cuachalalate". This is a medicinal tree widely used in Mexico for a variety of human affections like circulatory problems, stomach cancer, urinary disorders, gastrointestinal, lung and respiratory diseases, among others. It is an endemic Mexican tree, resinous and dioecious, inhabiting in regions with deciduous or sub-deciduous tropical forest [18]. Phytochemical studies of its bark have revealed the presence of a wide variety of compounds like masticadienonic acid, 3a-hidroximasticadienonic acid, alkyl phenolic acids, alkyl phenolic aldehydes, tirucallane-type triterpenes, anacardic acids and β-sitosterol, which exhibit different biological activities [19-21].
This report deals with the evaluation of the cytotoxic activity of an endophytic actinomycete isolated from A. adstringens and the chemical characterization of the most active compound to finally discuss its possible mechanism of action.
Plant material and collection site
Plant material was collected in Barranca Honda, municipality of Tlaltizapan, Morelos, Mexico (18°48'33.4476"N, 99°07'13.944"W). The first collection was in February 2013 (drought) by sampling stem vascular tissue and seeds from female trees and stem vascular tissue from male trees. There were no leaves on the trees at this season. The second collection was carried out in August 2013 (rainy), taking leaves, stem and seeds from the female trees as well as leaves and stems tissue from the male trees. Samples were identified by a specialist at the National Herbarium of Mexico as A. adstringens and incorporated as Herbarium specimens under numbers 1366483 for the female tree and 1366484 for the male tree.
Samples for microbiological analysis were sprayed with a 70% ethanol solution and stored in sterile bags at low temperature for transportation. Leaves and branches were also collected for plant identification.
Sample treatment and isolation of endophytes
A stem sample of A. adstringens was exhaustively washed in running water. Then cut out in a small piece and subject to a four-step surface sterilization procedure by sequential immersion (one minute each) in sterile water, 65% ethanol, 3.4% NaClO and sterile water (modified from Sanchez Fernández et al.). A 500 µl sample of the fourth wash was used to inoculate Nutrient agar to test for sterility of the sample surface. The sample was cut out into 0.3 cm2 pieces and placed in a Petri dish containing Nutrient agar Difco™ 213000 (Becton Dickinson, San Diego, CA, USA) for at least 60 days at 29°C. Complete isolation of the actinomycete was possible by sub-culturing the colonies in ISP-2 medium [22].
Molecular characterization of endophytic actinomycetes
The Actinomycete DNA was isolated using the ZR Fungal/Bacterial DNA MiniPrep™ (Zymo Research Corp, Irvine, CA, USA) commercial kit, following manufacturer's instructions. 16S ribosomal DNA sequences of the four actinomycetes were amplified using universal primers fwd5’ -CCGAATTCGTCGACAACAGAGTTTGATCATGGCTCAG and rev 3’ –CCCGGGATCCAAGCTTACGGCTACCTTGTTACGACTTTGTTACGACTT (Weisburg, et al.), following PCR amplification conditions were five min initial activation of Taq-DNA-Polymerase for five min at 94°C, followed by 30 cycles of 30 s denaturation at 94°C, annealing for 1 min at 56.5°C and extension for 2 min at 72°C, with a final extension step at 72°C for four min [14]. PCR product purification was accomplished using the commercial kit Wizard®SV Gel and PCR Clean-Up System (Promega Corporation, Madison, WI, USA) and sent to Laragen, Inc. (CA, USA) for sequencing.
The 16s rDNA sequence was deposited in GeneBank under the accession number KU579247. The sequence was compared using BLAST analysis with sequences in the existing 16S rDNA database at NCBI [23]. Phylogenetic tree of NF3 was built in Mega6 [24] using the Maximum Likelihood method based on the Jukes-Cantor model and a bootstrap value of 1000 and comparing with the strains that presented the closest proximity at the time of computation. The isolated actinomycete (NF3), is maintained in viable conditions at the UNAM-48/WFCC (Mexico City).
Microscopic characterization of NF3 strain
Erlenmeyer 250-mL flasks containing 50 mL ISP-2 liquid medium was inoculated with NF3 strain and incubated for 8 days at 29 °C, at 180 rpm. After that time, 200 μl of the culture were used to inoculate Petri dishes containing ISP-2 semi-solid culture media, gently dispersing with glass beads over the whole surface.
Microscopic slides were prepared from Gram stained colonies grown for 8 days in semi-solid ISP-2 medium. Preparations were observed in a bright field Primo Star Zeiss Microscope and stereoscopic Spencer microscope.
Colonies were also handled for visualization by Scanning Electron Microscopy (SEM) following a modified protocol from [25]. A colony was sampled with a 5 mm puncher and placed in 1.5 mL microcentrifuge tube containing 3% formaldehyde in phosphate-buffered saline (PBS) during 24h. Sample was washed with PBS and dehydrated using an ethanol gradient 30%, 50%, 70% 85% and 100 and, were placed in a Samdri-795 for drying at CO2 critical point and finally covered with gold in a Jeol ion sputter JFC-1100. Samples were observed with a JEOL JSM-6360LV Microscope.
Fermentation and purification of the major compound from strain NF3
A 14-L New Brunswick bioreactor fitted with Rushton Turbines and air diffusion was used, containing 10 liters ISP-2 medium. The sterilized medium was inoculated with a 10% v/v (3 days inoculum) of the strain NF3 and the fermentation was conducted at 60 rpm and 0.5 vvm for 8 days at 29°C. After fermentation, the culture media was centrifuged at 10,000 x g for 10 min. Supernatant was exhaustively extracted by partition and Biomass by maceration, both with ethyl acetate (EtOAc) until no color appeared in the extraction solvent. The organic solvent extracts were evaporated under reduced pressure in a Büchi rotary evaporator R-215 coupled to the vacuum pump V-700 and mixed, yielding approximately 1 g of crude extract, which was subjected to a 77 cm long open column chromatography, packed with 169 g of silica gel 60 (70-230 mesh) from Merck-Millipore (Darmstadt, Germany). The column was eluted with a gradient of n-hexane/CH2Cl2 (100:0 to 0:100) and CH2Cl2/MeOH (99.9:0.1 to 0:100) resulting in 14 primary fractions.
The more active fraction was analyzed by HPLC/UV/ESI-MS in an Agilent 1200 Binary SL pump (Santa Clara, CA, USA) coupled with an ESI-MS Mass Spectrometer Esquire 600 (Bruker, Billerica, MA, USA) and a Waters 2996 PDA detector (Milford, MA, USA). For High-performance liquid chromatography (HPLC), a Synergi 4u Polar RP 80A 150 X 2.0 mm column (Phenomenex, Torrance, CA, USA) was used with MeOH:H2O (80:20, v/v) to 100% MeOH at 0.2 mL/min flow. The major compound was purified in a Millipore preparative TLC Silica gel 60 F254 Glass plates 20 X 20 cm (Merck-Millipore, Darmstadt, Germany), being the elution system CH2Cl2/MeOH (99:1), yielding 20 mg of an orange powder. Nuclear Magnetic Resonance spectrometry (NMR) was done in a Bruker 400 MHz Spectrometer using deuterated chloroform as solvent. A Xevo G2-XS QTOF mass spectrometer (Waters, Milford, MA, USA) was used to compare the fragmentation pattern of the isolated compound with a commercial standard of Steffimycin B from Enzo Life Sciences ALX-380-220-M001 (Ann Arbor, MI, USA).
Cell lines culture
Breast adenocarcinoma (MCF-7 ATCC HTB-22), human cervical carcinoma (HeLa ATCC CCL-2) and immortal keratinocytes (HaCaT) were donated by Centro Médico Nacional Siglo XXI, México, and authenticated by Instituto Nacional de Medicina Genómica, México. MCF-7 and HaCaT cell lines were cultured in RPMI-1640 medium supplemented with L-glutamine Caisson RPL03 (Caisson Laboratories, East Smithfield, UT, USA), 10% (v/v) fetal bovine serum and 1% penicillin/streptomycin. HeLa cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with high glucose, L-glutamine, and sodium pyruvate Caisson DML10, 10% (v/v) fetal bovine serum and 1% (w/v) penicillin/streptomycin. Cells were incubated at 37°C with 98% humidity and a 5% CO2 gas environment.
For the pure compound evaluation, different breast cancer cell lines including triple negative were also tested, with HCC1806 ATCC crl-2335, HCC1937 ATCC CRL-2336, DU4475 ATCC HTB-123 and MBCD5 (kindly donated from Instituto Nacional de Ciencias Médicas y Nutrición), all of them cultured in RPMI-1640 medium as described above.
In Vitro Cytotoxicity Assay
Cell survival and proliferation were determined by a Cell Counting Kit-8 WST-8 (Dojindo Molecular Technologies, Inc., Rockville, MD, USA) in a 96-well plate containing 5000 cells per well and incubated for 24hr. Crude extract, fractions and the pure compound were added from DMSO-diluted stocks to achieve different concentrations of the extract per well (0.1 to 100 μg/mL). In all cytotoxic assays, DMSO was tested as the diluent control (vehicle); sterile media was used as a negative control and doxorubicin at the same concentrations was tested as a positive control. After 48 h incubation with treatments, 10 μL of WST-8 was added to each well and 2h later, the absorbance was measured at 450 nm in a Thermo Scientific Multiscan™ FC Microplate spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). When the final concentration of DMSO was higher than 0.5% v/v (only in concentrations at 100 μg/mL), the activity for DMSO was subtracted from the result, being 20% mortality for MCF7 and 22% for HeLa cells, not DMSO mortality was detected for HaCaT cells at any concentration. A calibration curve was prepared with known cell numbers and the percent of mortality calculated based on the number of untreated cells. IC50 values were determined from the plot’s percent mortality against extract concentration and calculated by logarithmic regression analysis. Results are representative of three individual experiments.
Detection of the apoptotic cells by flow cytometry
A million of MCF7 cells were incubated for 24h and treated with the purified steffimycin B (StefB) at the IC50 value for 12, 24 and 48h. Doxurubicin treated cells at IC50 value for 48h were tested as positive apoptotic control and vehicle and not treated cells were evaluated at 48h as negative controls. Cells were harvested using a versene solution for cell dissociation and washed with PBS, without any previous wash. Cell pellet were treated with the APC Annexin V Apoptosis detection kit with 7-AAD Cat 640930 (BioLegend, San Diego, CA, USA) according to the manufacturer’s instructions. Samples were analyzed in an Attune acoustic focusing cytometer measuring 10,000 cells using the Attune cytometric software (Thermo Fisher Scientific, Waltham, MA, USA).
Proliferation by Western blot analysis
MCF7 cells were incubated and treated as in the flow cytometry analysis. Cells were resuspended in 75 μl lysis buffer (50 mM TRis-HCl pH 8; 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1mM EDTA; 1 mM Na3VO4) containing 40 μL/mL of protease inhibitors cocktail (Roche Applied Science, Mannheim, Germany) and 10 μL/mL of phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO, USA). Cells were kept under stirring for 20 min at 4 °C, then centrifuged at 14,000 rpm for 3 min. Supernatant was recovered and stored at -20 °C until use. Protein concentration was calculated using the DC Protein Assay kit (Bio-Rad Laboratories, Hercules, CA, USA). Protein (30 μg/well) was separated on 12% SDS-PAGE and electro-transferred onto Immobilon-PSQ (polyvinylidene fluoride) membrane (Millipore, Billerica, MA, USA). Membranes were blocked with 3% skimmed milk powder in Tris-buffered saline (TBS) for 30 min and incubated for 24h with Anti-PCNA antibody [EPR3821] ab92552 (Abcam, Cambridge, UK) diluted 1:1000 at 4°C. As an internal control, a rabbit anti-GAPDH GTX100118 (Genetex Inc., Irving, CA, USA) was used. After incubation with the first antibody, membrane was washed three times with TBS-Tween 5% and then incubated for 1h at room temperature with the goat anti-rabbit IgG-HRP sc-2004 Secondary Antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 1:15,000 dilution. Chemiluminescent bands were detected using the kit Trident ECL plus GTX400006 (Genetex Inc., Irving, CA, USA) and images were obtained in a Carestream Kodak BIOMAX Light Film CAT 178 8207 (Sigma-Aldrich, St. Louis, MO, USA).
Evaluation of senescence and morphological changes
24-well plates containing 10,000 MCF7 cells per well were treated for flow cytometry assay. To detect senescence cells, a β-Galactosidase Staining Kit (Cell Signaling Technology, INC., Danvers, MA, USA) was used following manufacturer's instructions. Plates were observed in an Olympus IX-71 microscope (Olympus Corporation, Tokio, Japón) adapted with an Olympus LUCPlanFL N 20x (N.A. 0.45) objective. Images were acquired with an Evolution VF camera (Media Cybernetics, Rockville, MD, USA).
Isolation and identification of the NF3 strain
The NF3 strain was isolated from a small piece of a female stem of A adstringens incubated in nutrient agar medium. The strain presented small mycelium at X1000 magnification and irregular shape colonies at X250, a morphology characteristic of actinomycetes. The 16S ribosomal DNA from NF3 strain with 1272 bp (base pairs) showed 99% identity with different strains of Streptomyces scabrisporus. The phylogenetic tree of the NF3 (Figure 1), exhibited this strain in a clade where all the S. scabrisporus reported up today are included, with a high value of bootstrap, supporting its identity with this species. Its identification was corroborated as S. scabrisporus via 16S rRNA, gyrB, rpoB and recG genes [26].
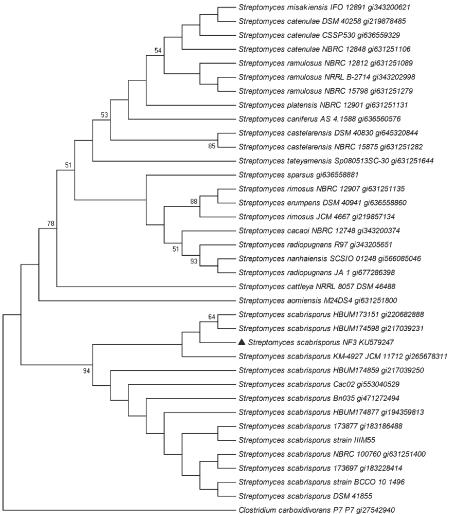
Figure 1. Phylogenetic tree of the NF3 strain using the maximum likelihood method based on the Jukes-Cantor model. Bootstrap values were calculated from 1,000 re-samples with values over 50% shown at the respective node.
Figure 2 shows the S. scabrisporus NF3 strain growing as soft pink colonies in rough aggregates, changing to darker colonies with white spores that appeared at the time of incubation, as well as producing a red pigmentation in agar. Gram-positive mycelia formation was observed under optic microscopy while in SEM micrographs lineal spore chains presenting some areas with free spores were found.
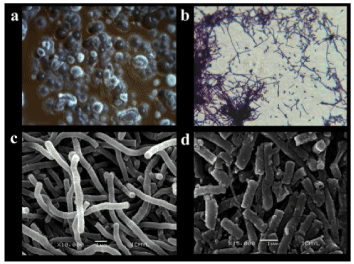
Figure 2. Micrographs of S. scabrisporus at: a) stereoscopic microscope (X250); b) Optical microscope (X1,000); c) SEM micrograph (X10,000) of lineal mycelia; d) SEM micrograph (X15,000) of spore formation.
Identification of the major bioactive compound of S. scabrisporus NF3
The S. scabrisporus NF3 crude extract obtained from a 10-liter bioreactor was evaluated against MCF7, HeLa and HaCat cell lines (Table 1) finding a high cytotoxic activity when compared with the positive control Doxorubicin.
Table 1. Cytotoxic effect of NF3 crude extract grown in ISP-2 medium and the positive control Doxorubicin, against different cell lines
EXTRACTS |
IC50 (μg/mL) |
MCF7 |
HeLa |
HaCaT |
NF3 EtOAc |
0.96 |
1.98 |
2.7 |
Doxorubicin |
0.72 |
2.4 |
2.5 |
Fractions obtained in the open chromatography column were also evaluated, finding that 9 out of 14 fractions showed high activity against the same three cell lines (Table 2).
Table 2. Cytotoxic effect of the fractions obtained from an open chromatography column from the S. scabrisporus NF3 strain
Fractions* |
% Mortality (at 100 μg/mL) |
|
MCF7 |
HeLa |
HaCaT |
Total fraction (mg) |
F1 |
0 |
8.9 ± 1.7 |
45.7 ± 0.9 |
16.0 |
F2 |
0 |
0.7 ± 1.0 |
34.8 ± 1.3 |
58.5 |
F3 |
0 |
7.1 ± 2.0 |
35.1 ± 2.8 |
25.1 |
F4 |
69.4 ± 0.2 |
66.9 ± 1.2 |
44.5 ± 0.9 |
197.6 |
F5 |
80.8 ± 0.4 |
78.3 ± 0.6 |
61.4 ± 0.4 |
20.7 |
F6 |
77.5 ± 1.3 |
78.8 ± 0.1 |
61.6 ± 0.2 |
148.7 |
F7 |
79.1 ± 0.6 |
78.7 ± 0.5 |
60.0 ± 0.5 |
132.1 |
F8 |
79.1 ± 0.8 |
79.0 ± 0.5 |
60.9 ± 0.6 |
116.7 |
F9 |
80.0 ± 0.2 |
100.0 ± 0.3 |
61.3 ± 0.7 |
59.0 |
F10 |
0 |
32.9 ± 4.9 |
3.5 ± 1.0 |
46.9 |
F11 |
0 |
9.1 ± 1.3 |
0 |
55.6 |
F12 |
97.6 ± 0.3 |
94.2 ± 2.2 |
58.6 ± 0.3 |
109.1 |
F13 |
97.0 ± 0.4 |
98.3 ± 0.1 |
58.3 ± 0.4 |
19.8 |
F14 |
89.4 ± 1.1 |
69.9 ± 2.3 |
55.3 ± 0.3 |
60.7 |
*Fractions 1 to 8 were diluted in DMSO, and DMSO activity was subtracted to the final result. Fractions 9 to 14 were diluted in water.
Because of its high activity against the cancer cell lines and low activity against keratinocytes, fraction 4 was chosen for subsequent purification. Besides, 197.6 mg were obtained from this fraction. The HPLC/UV/ESI-MS analysis showed 10 peaks in the chromatogram, being the peak 8 the major compound (Figure 3a) and the other compounds in that fraction seemed to be closely related because of its yellow color and proximity of mass profiles (data not shown). The mass and the UV spectra of this compound can be seen in Figure 3b and 3c, respectively. These data and the 1H and 13C NMR spectra supported StefB as the active molecule (Figure 3d). The ESI-MS results are as follow: m/z 587.5 [M-H]; UV in methanol: lmax at 235.5 nm. 1H NMR (CDCL3, 400 MHz): δ 12.91 (s, 1H), 12.0 (s, 1H), 8.29 (s, 1H), 7.35 (d, J = 2.8 Hz, 1H), 6.64 (d, J = 2.4 Hz, 1H), 5.54 (s, 1H), 5.12 (d, J = 2.4 Hz, 1H), 3.89 (s, 3H), 3.72 (m, 1H), 3.69 (d, J = 2.5 Hz, 1H), 3.61 (m, 1H), 3.56 (s, 1H), 3.52 (s, 3H), 3.51 (s, 3H), 3.49 (s, 3H), 3.47 (dd, J = 3.6, 1H), 3.00 (t, 9.4 Hz, 1H) , 2.34 (d, J = 8.8 Hz, 1H), 1.45 (s, 3H), 1.33 (s, 3H); 13C NMR (CDCL3, 100MHz): δ 198.94, 190.64, 180.41, 167.5, 165.85, 162.15, 135.53, 135.07, 133.55, 133.52, 118.77, 117.59, 110.18, 109.27, 106.75, 100.6, 85.67, 83.3, 80.46, 76.68, 71.95, 71.27, 69.00, 60.94, 59.98, 58.85, 56.29, 22.9, 17.95. Fragmentation patterns of the compound isolated in this study, named 4D along the purification steps (Figure 3e), and a commercial standard of StefB (Figure 3f) were acquired, obtaining similar results.
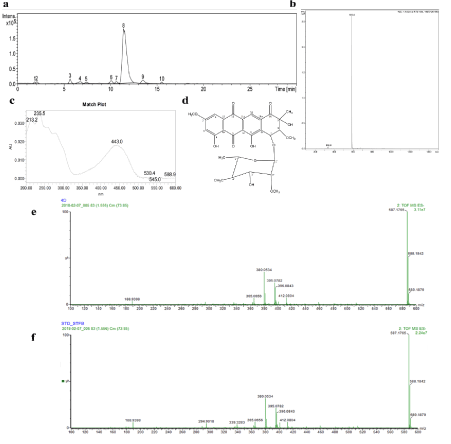
Figure 3. Chemical characterization of fraction 4 and structure confirmation of StefB. a) HPLC chromatogram; b) Mass spectra of compound 8; c) UV spectra of compound 8; d) Chemical structure of major compound Stef B; e) Fragmentation pattern of the major compound 4D identified as StefB; f) Fragmentation pattern of commercial standard of StefB.
Cytotoxic activity of StefB and its effect on MCF7 cells
Cytotoxic activity of the pure compound was tested against different cell lines, including three triple negative breast cancer cell lines (HCC1806, HCC1937 and DU4475). As shown in Table 3, StefB affected MCF7, DU4475, HCC1806 and MBCD5 cell lines, with MCF7 as the most sensitive one. As seen in the same table, although the IC50 values were higher than doxorubicin, this commercial compound also affected normal keratinocytes.
Table 3. IC50 values of the cytotoxic activity of StefB in different cell lines
CELL LINE |
IC50 μM |
StefB |
Doxorrubicin |
HeLa |
71.90 |
4.34 |
MCF7 |
2.56 |
1.32 |
HaCaT |
156.00 |
4.67 |
HCC1806 |
18.20 |
0.07 |
HCC1937 |
456.00 |
0.45 |
MBCD5 |
25.70 |
1.59 |
DU4475 |
4.81 |
<0.09 |
To elucidate the possible effects of StefB, treated MCF7 cells were analyzed by flow cytometry and Western blot. Figure 4a shows the presence of apoptotic cells, after treating the cell line with 2.56 µM StefB, with 48 h exposure as the time to get the maximum value. Furthermore, the presence of StefB, negatively affected cell proliferation, as judged by a Figure 4 western blot analysis of the PCNA protein (Figure 4b). Finally, strong morphological changes were microscopically detected after StefB treatment with the presence of senescence cells (stained in blue), with a maximum effect at 48 h treatment (Figure 4c).
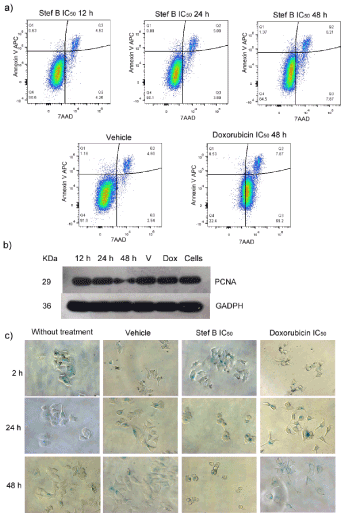
Figure 4. Effect of StefB on MCF7 cells. a) Flow cytometry analysis evaluating apoptosis and necrosis; b) Western Blot analysis of the expression of the proliferation marker PCNA (find at 29 KDa) using glyceraldehyde-3-phosphate dehydrogenase (GADPH) as positive control (at 36 KDa); c) Micrographs showing morphological changes of the cells stained with β-galactosidase.
In this study, an actinomycete (NF3 strain) isolated from a female stem from the medicinal tree A. adstringens was selected by their ability to produce secondary metabolites, property frequently found in endophytic actinomycetes [27-29]. Its molecular identification highlighted NF3 strain as S. scabrisporus, a species never reported as a plant endophyte.
The first report on S. scabrisporus KM-4927T (=JCM 11712T = NRRL B-24202T) described this microbe as a gray aerial cell mass producer with spiral spore chains and a rough spore surface [30] unable to produce soluble pigments on ISP-2 medium and other solid media. A recent study also reported another strain of S. scabrisporus with spiral aerial mycelium and unable to produce soluble pigments when grown in ISP-2 medium [30,31]. Contrary to the S. scabrisporus NF3 strain, those strains were isolated from soil. Interestingly, the NF3 strain produced pigments in different media, including ISP-2. Regarding its morphology, SEM micrographs showed lineal spore chains formation instead of the multiple-spiral spore chains reported for other S. scabrisporus strains [30], further supporting differences between these microbes. These differences could be related to the activation of pathways used in the symbiotic relationship between NF3 strain, and A. adstringens.
The genome sequence of various S. scabrisporus strains, have been already released. Their analysis suggested variations between KM-4927T and DSM 41855 strains with changes in some of their polyketide biosynthetic genes [32]. Furthermore, a recent comparison made by our research group between the strains DSM 41855 and NF3, suggested important differences in their putative gene clusters encoding for secondary metabolites production [33]. Finally, the presence of 2 clades in the S. scabrisporus phylogenetic tree, with more than 60% bootstrap value, may support significant dissimilarities within this species.
Chromatography and HPLC/UV/ESI-MS analysis of the S scabrisporus NF3 organic extracts identified the major fraction with cytotoxic activity as StefB, a bioactive anthracycline initially reported in 1974 in Streptomyces elgreteus [34]. Similar fragmentation patterns of the isolated compound with those of a commercial standard of StefB, supported further the authenticity of the cytotoxic compound.
Other S. scabrisporus strains have been reported to produce bioactive compound like the macrocyclic hitachimycin [32], the polyketide okilactomycin [35] and the polyether ionophore alborixin [36], all of them colorless compounds with antibiotic and cytotoxic activities. In addition to StefB, S. scabrisporus NF3 was found to produce two uncharacterized bioactive pigments (red and yellow), not previously described for this species.
The activity of StefB has been reported against Gram-positive bacteria with low inhibitory effect on L.1210 mouse leukemia cells [34]. Antimalarial and antimycobacterial activities have also been reported for this molecule, as well as against both malignant (KB, NCI-H187) and non-malignant (Vero) cells [37,38]. On the contrary, antifungal activity against fourteen human pathogenic fungi was not detected [34].
Studies to improve the activity and solubility of this molecule either via chemical, microbiological or molecular biology have also been performed by different groups [39-41]. Furthermore, isolation, characterization and heterologous expression of the StefB gene cluster, was also explored [42].
When StefB was tested over different cell lines not previously evaluated, this compound affected from major to minor scale the breast adenocarcinoma MCF7, the triple breast cancer cell lines DU4475, HCC1806, MBCD5 and Hela, showing a high IC50 value against the non-tumor cell line HaCaT. Among them, the triple negative HCC1806 and DU4475 have gained importance in the last years mainly due to its resistance to targeted treatments. When compared to doxorubicin, StefB resulted less aggressive or toxic than the above anticancer compound, making it an attractive drug for further studies. Doxorubicin was selected as positive control in this study since also belongs to the anthracycline’s family of bioactive compounds [43].
Regarding to its mechanism of action, StefB binds to double-stranded DNA as showed by difference spectroscopy and an increase of the DNA thermal stability in the presence of the anthracycline [44]. Furthermore, the molecular and crystal structure of StefB has been described and used for modeling the intercalative binding with DNA, finding that StefB has a higher CpG sequence specificity over the TpA step, similar to that of daunorubicin and nogalamycin [45].
The behavior of the tumor cells to the presence of StefB has not been characterized. In our study we evaluated this response in the MCF7 cell line, as this was the most sensitive to the StefB effect. In this line, a decrease of cell proliferation via PCNA expression was observed with the presence of apoptotic cells and strong morphological changes with the presence of senescence cells that increased with the exposure time to StefB. Although apoptosis seems to be the cell death pathway as occurs with doxorubicin, the extremely morphological changes caused by StefB makes necessary to perform additional studies to know why these structural modifications occur in the cells and their subsequent implications.
In conclusion, an actinomycete with potential to produce bioactive compounds was isolated from Amphypterigium adstringens. This strain identified as S. scabrisporus was selected by a bioassay-guided study showing a strong cytotoxic potential against various cancer cell lines (breast and cervical carcinoma). A major compound purified from crude extracts of this strain and responsible of the biological activity was identified as StefB. This compound, formerly known for its different biological activities, strongly affected different breast cancer cell lines, including triple negative ones, with low toxicity against keratinocytes, making it a promissory drug for breast cancer treatment.
Karol Rodríguez-Peña is a doctoral student from Programa de Doctorado en Ciencias Biomédicas, UNAM. She was recipient of the fellowship 161183 from CONACYT, México. We are grateful to G. Sánchez-Ruiz, R. González-Moguel, L. Tovilla-Coutiño, C. Aguilar-Zacarías and B. Ruiz-Villafán for their valuable participation in this work. We thank Y. Hornelas-Orozco from ICMYL-UNAM, for the scanning electronic micrographs, K. Bermudez-Torres from CEPROBI-IPN for her advice in the collection of the plant samples, R. Medina-Lemos from Instituto de Biología-UNAM, for plant identification, Abel Blancas for the Biorreactor facilities, M.A. Ortiz-Jiménez for the actinomycete preservation and M. Tapia for the microscopic images.
The authors declare that they have no conflict of interest
This article does not contain any studies with human participants performed by any of the authors
KRP, SS, MLMR, LRZ and RRS contributed equally to conception and design of the project, data analysis and interpretation. KRP and MTU contributed to acquisition data. All authors revised the manuscript and gave the final approval of it and agree to act as guarantors of the work.
This work was supported by the DGAPA, PAPIIT grant UNAM IN-202216 and the NUATEI program from Instituto de Investigaciones Biomédicas, UNAM. We thank the support of CONACYT project number INFR-2017-01 279880, which allowed the acquisition of an HPLC masses.
- Gravitz H (2012) Turning a new phage. Nat Med 18: 1318-1320. [Crossref]
- Demain AL, Sanchez S (2015) The need for new antibiotics: Antibiotics: Current Innovations and Future Trends 1st Eds. Caister Academic Press, 65-82.
- Schulz B, Boyle C (2005) The endophytic continuum. Mycol Res 109: 661–686. [Crossref]
- Guzmán-Trampe S, Rodríguez-Peña K, Espinosa-Gómez A, Sánchez-Fernández RE, Macías-Rubalcava ML, Flores-Cotera LB, Sánchez S (2015) Endophytes as a potential source of new antibiotics: Antibiotics: Current Innovations and Future Trends 1st Eds. Caister Academic Press, 175-204.
- Stierle A, Strobel G, Stierle D (1993) Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of pacific yew. Science 260: 214–216. [Crossref]
- Flores-Bustamante ZR, Rivera-Orduña FN, Martínez-Cárdenas A, Flores-Cotera LB (2010) Microbial paclitaxel: advances and perspectives. J Antibiot 63: 460–467. [Crossref].
- Rajendran L, Rajagopal K, Subbarayan K, Ulagappan K, Sampath A, et al. (2013) Efficiency of fungal taxol on human liver carcinoma cell lines. Am J Res Commun 1: 112–121.
- Yang Y, Zhao H, Barrero RA, Zhang B, Sun G, et al. (2014) Genome sequencing and analysis of the paclitaxel-producing endophytic fungus Penicillium aurantiogriseum NRRL 62431. BMC Genomics 15: 1-14. [Crossref]
- Nadeem M, Ram M, Alam P, Ahmad MM, Mohammad A, et al. (2012) Fusarium solani, P1, a new endophytic podophyllotoxin-producing fungus from roots of Podophyllum hexandrum. African J Microbiol Res 6: 2493–2499.
- Shweta S, Gurumurthy BR, Ravikanth G, Ramanan US, Shivanna MB (2013) Endophytic fungi from Miquelia dentata Bedd., produce the anti-cancer alkaloid, camptothecine. Phytomedicine 20: 337–342. [Crossref]
- Kharwar RN, Mishra A, Gond SK, Stierle A, Stierle D (2011) Anticancer compounds derived from fungal endophytes : their importance and future challenges. Nat Prod Rep 28: 1208-1228. [Crossref]
- Martinez-Klimova E, Rodríguez-Peña K, Sánchez S (2017) Endophytes as sources of antibiotics. Biochem Pharmacol 134: 1-17. [Crossref]
- Sánchez-Fernández RE, Diaz D, Duarte G, Lappe-Oliveras P, Sánchez S, et al. (2015) Antifungal volatile organic compounds from the endophyte Nodulisporium sp. strain GS4d2II1a : a qualitative change in the intraspecific and interspecific interactions with Pythium aphanidermatum. Microb Ecol 71: 347–364. [Crossref]
- Guzmán-Trampe S, Lemus D, Jiménez O, Ruiz-Villafán B, García-Carrancá A, et al. (2015). Evaluation of the potential bioactivity of an endophytic bacterium isolated from Magnolia dealbata Zucc. Intl J Curr Microbiol Appl Sci 4: 515–525.
- Zhang G, Sun S, Zhu T, Lin Z, Gu J, et al. (2011) Antiviral isoindolone derivatives from an endophytic fungus Emericella sp. associated with Aegiceras corniculatum. Phytochemistry 72: 1436–1442. [Crossref]
- Doroghazi JR, Metcalf WW (2013) Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC genomics 14: 611–624. [Crossref]
- Golinska P, Wypij M, Agarkar G, Rathod D, Dahm H, et al. (2015) Endophytic actinobacteria of medicinal plants: diversity and bioactivity. Ant van Leeuw 108: 267-289. [Crossref]
- Cuevas X (2005) A revision of the genus Amphipterygium (Julianiaceae). Ibugana 13: 27-48.
- Mata R, Calzada F, Navarrete A, Del Rio F, Delgado G (1991) Long-chain phenols from the bark of Amphypterygium adstringens. J Ethnopharmacol 34: 147-154.
- Oviedo-Chávez I, Ramı́rez-Apan T, Soto-Hernández M, Martı́nez-Vázquez M (2004) Principles of the bark of Amphipterygium adstringens (Julianaceae) with anti-inflammatory activity. Phytomedicine 11: 436–445. [Crossref]
- Makino M, Motegi T, Fujimoto Y (2004). Tirucallane-type triterpenes from Juliania adstringens. Phytochemistry 65: 891–896. [Crossref]
- Shirling E, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Sys Bacteriol 16: 313–340.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [Crossref]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [Crossref]
- Bozzola JJ, Russell LD (1992) Electron Microscopy, principles and techniques for biologists, 1st edn. Jones and Burtlett Publishers USA.
- Ceapa C, Vázquez-Hernández M, Rodríguez-Luna SD, Cruz-Vázquez AP, Jiménez-Suárez V, et al. (2018) Genome mining of Streptomyces scabrisporus NF3 reveals symbiotic features including genes related to plant interactions. PLos One 13: e0192618. [Crossref]
- Golinska P, Wypij M, Agarkar G, Rathod D, Dahm H, et al. (2015) Endophytic actinobacteria of medicinal plants: diversity and bioactivity. Ant van Leeuw 108: 267-289. [Crossref]
- Wu Y, Lu C, Qian X, Huang Y, Shen Y (2009) Diversities within genotypes, bioactivity and biosynthetic genes of endophytic actinomycetes isolated from three pharmaceutical plants. Curr Microb 59: 475–482. [Crossref]
- Janso JE, Carter GT (2010) Biosynthetic potential of phylogenetically unique endophytic actinomycetes from tropical plants. Appl Environ Microbiol 76: 4377–4386. [Crossref]
- Ping X, Takahashi Y, Seino A, Iwai Y, Omura S (2004) Streptomyces scabrisporus sp. nov. Int J Syst Evol Microbiol 54:577–581. [Crossref]
- Charousova I, Medo J, Halenárová E, Javorekova S (2017) Antimicrobial and enzymatic activity of actinomycetes isolated from soils of coastal islands. J Adv Pharm Technol Res 8: 46-51. [Crossref]
- Kudo F, Kawamura K, Uchino A, Miyanaga A, Numakura M, et al. (2015) Genome mining of the hitachimycin biosynthetic gene cluster: Involvement of a phenylalanine-2,3-aminomutase in biosynthesis. Chembiochem 16: 909-914. [Crossref]
- Vazquez-Hernandez M, Ceapa CD, Rodríguez-Luna SD, Rodríguez-Sanoja R, Sánchez S (2017) Draft genome sequence of Streptomyces scabrisporus NF3, an endophyte isolated from Amphipterygium adstringens. Genome Announce 5: 1-2. [Crossref]
- Brodasky TF, Reusser F (1974) Steffimycin B, a new member of the steffimycin family: isolation and characterization. J Antibiot 27: 809-813. [Crossref]
- Zhang C, Ondeyka JG, Zink DL, Basilio A, Vicente F, et al. (2009) Discovery of okilactomycin and congeners from Streptomyces scabrisporus by antisense differential sensitivity assay targeting ribosomal protein S4. J Antibiot 62: 55-61. [Crossref]
- Manzoor A, Wani A, Qazi PH, Rehman S, Mushtaq S, et al. (2016) Isolation and characterization of alborixin from Streptomyces scabrisporus: A potent cytotoxic agent against human colon (HCT-116) cancer cells. Chem Biol Interact 256:198-208.
- Intaraudom C, Bunbamrung N, Dramae A, Danwisetkanjana K, Rachtawee P, et al (2015) Antimalarial and antimycobacterial agents from Streptomyces sp BCC27095. Tetrahedron Lett 56: 6875-6877.
- Olano C, Abdelfattah MS, Gullón S, Brana AF, Rohr J, et al. (2008) Glycosylated derivatives of steffimycin: Insights into the role of the sugar moieties for the biological activity. Chembiochem 9: 624-633. [Crossref]
- Wiley PF, Elrod DW, Slavicek JM, Marshall VP (1980) Microbial conversion of steffimycin and steffimycin B to 10-dihydrosteffimycin and 10-dihydrosteffimycin B. J Antibiot 33: 819-823. [Crossref]
- Kelly RC, Schletter I, Koert JM, MacKellar FA, Wiley P (1977) Structures of steffimycin and steffimycin B. J Org Chem 42: 3591-3596.
- Wiley PF, Elrod DW, Harper D (1987) Chemical modification of steffimycin B. J Antibiot 41: 343-351. [Crossref]
- Gullon S, Olano G, Abdelfattah MS, Brana AF, Rohr J, et al (2006) Isolation, characterization, and heterologous expression of the biosynthesis gene cluster of the antitumor anthracycline steffimycin. Appl Environ Microbiol 72: 4172-4183. [Crossref]
- Rabbani A, Finn RM, Ausió J (2004) The anthracycline antibiotics: antitumor drugs that alter chromatin structure. BioEssays 27: 50-56. [Crossref]
- Reusser F (1974) Steffimycin B, a DNA binding agent. Biochim Biophys Acta 383: 266-273.
- Sriram M, Liaw YC, Gao Y-G, Wang H-J (1991) Molecular structure of antitumor drug steffimycin and modeling of its binding to DNA. J Biomol Struct Dyn 9:251-269. [Crossref]




