Abstract
Small Ubiquitin-related Modifier (SUMO) is a member of the ubiquitin-like protein family. These proteins, which are typically 70-100 amino acid residues in length each become covalently conjugated to lysine side chains in multiple conjugation targets altering the function, subcellular localization, and turnover of the targets. Like other members of the family, SUMO is conserved throughout the eukaryotic domain. Analysis of Drosophila SUMO combining biochemical approaches with the powerful genetic and reverse genetic approaches available in the Drosophila system has revealed multiple roles for SUMO conjugation in metazoan development. Furthermore, studies in mammalian systems show that most of these functions are conserved in mammals. These include roles in signal transduction via pathways such as the Ras/MAPK, Decapentaplegic (Dpp), Jun N-terminal Kinse (JNK), and Toll signaling pathways. Studies of the role of SUMO in Toll signaling have revealed roles in modulating the innate immune response. In addition, SUMO regulates developmental pattern formation by modulating the activities of transcriptional regulatory proteins such as the Polycomb group protein Scm. By converting linear polypeptides into branched polymers, SUMO may help to increase protein functional diversity.
Keywords
SUMO, Ubc9, ubiquitin-like proteins, Ras/MAPK signaling, Dpp, Medea, Drosophila development, Scm, Groucho, Bicoid, Spalt, Toll, Innate Immunity
The SUMO Pathway
The ubiquitin-like protein Small Ubiquitin-related Modifier (SUMO) becomes covalently conjugated to a large variety of target proteins [1-3]. SUMOylation is a reversible protein modification and can thus reversibly alter protein activity. In this respect, SUMOylation is akin to other reversible protein modifications such as phosphorylation, acetylation, and methylation. However, in the case of SUMOylation, the much larger size of the modifying group increases the potential for molecular recognition.
SUMOylation is a multistep process (Figure 1). First, an actvating enzyme (SAE1/SAE2) reacts with ATP resulting in the adenylation of an active site cysteine residue. The adenylate group is then displaced by the C-terminal carboxyl group of SUMO leading to the formation of a thioester linkage. SUMO is then handed off to an active site cysteine residue in the SUMO conjugating enzyme (Ubc9) before being transferred to the amino group of a lysine side chain in a final target protein forming an isopeptide linkage between the lysine and the C-terminus of SUMO. Unlike ubiquitylation, in which the final transfer to the target protein absolutely requires a ligase to assist in target recognition, SUMOylation does not have an absolute requirement for a ligase. Nonetheless, ligases, including the PIAS family proteins, RanBP2, and Pc2 [2, 4-6] often assist in the final transfer (Figure 1). Lysine residues that are targeted for SUMOylation frequently fall within a sequence resembling ΨKXE (Ψ is any hydrophobic amino acid and X is any amino acid) [7]. SUMO modification can be reversed by SUMO deconjugates such as Ulp1, Ulp2, or related proteins, which catalyse the hydrolysis of the isopeptide linkage between SUMO and the lysine side chain in the target protein [8].
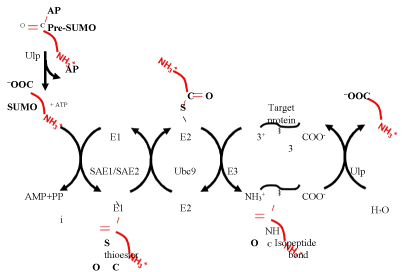
Figure 1. SUMO conjugation and deconjugation. SUMO is initially synthesized as a pre-protein with a C-terminal extension. In Drosophila, this extension is two amino acids long and has the sequence Ala-Pro (AP). The AP is cleaved off by a Ulp family protease to generate mature SUMO. SUMO is then attached to a target protein via a three-step pathway involving the E1 activating enzyme SAE1/SAE2, the E2 conjugating enzyme Ubc9, and a ligation step, which may or may not require an E3 enzyme. The resulting isopeptide bond between the target protein and the C-terminus of SUMO can be hydrolyzed by a Ulp family protease.
SUMO-modified proteins are able to interact non-covalently with other proteins through SUMO interaction motifs (SIMs). These motifs possess a hydrophobic core with the consensus sequence V/I-V/I-X-V/I (X is any amino acid) [9,10]. The SIM forms a b strand that interacts with the β2 strand of SUMO in either a parallel or anti-parallel orientation [11,12]. Serine and threonine residues adjacent to the SIM hydrophobic core can be phosphorylated, and the phosphate group forms a salt bridge to a conserved lysine residue within SUMO [10].
SUMO in Drosophila and Mammalian Development
The remainder of this review will focus on a few of the many roles of SUMO in regulating signal transduction and development in Drosophila melanogaster. This will include a discussion of the role of SUMO in the Ras/MAPK, Decapentaplegic (Dpp), and Jun N-terminal Kinase (JNK) signalling pathways, as well as a discussion of the role of SUMO in Polycomb group mediated repression. In addition, we will review the evidence that SUMO regulates signalling through the Toll like receptors, and thus the innate immune response. Furthermore, we will present evidence showing that, in many cases, these functions in Drosophila are conserved in mammals.
Ras/MAPK signalling
The Ras/MAPK signal transduction pathway (Figure 2) is required to pattern the follicle cell epithelium during egg chamber development [13]. This requires the secretion of the TGF-a-like protein Gurken from the presumptive dorsal side of the oocyte, and the binding of Gurken to the Torpedo receptor tyrosine kinase (RTK) in the membranes of the overlying follicle cells. Subsequent dimerization and cytoplasmic autophosphorylation of Torpedo leads to the formation of a docking site for the adaptor protein DRK [14-16]. DRK, in turn, recruits the GTP exchange factor Son of Sevenless (SoS) for Ras activation through the exchange of GDP for GTP in the membrane tethered Ras protein [17]. Ras then stimulates a phosphorylation cascade involving the sequential activation of three Ser/Thr kinases, Raf, MEK, and MAPK [18-20], thus triggering the adoption of a dorsal follicle cell fate. These follicle cells then secrete dorsal eggshell structures such as the pair of dorsal appendages that act as respiratory filaments [21-23].
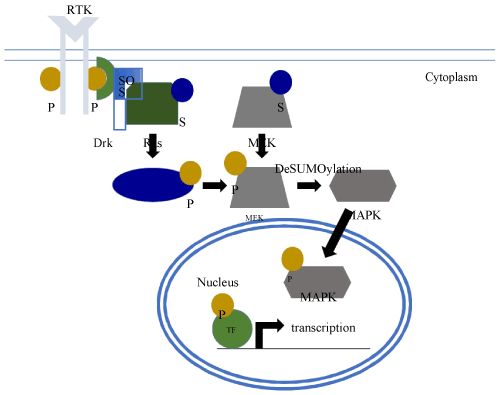
Figure 2. Regulation of RAS/MAPK by SUMO. Upon activation of a receptor tyrosine kinase (RTK), SUMO (S) modification of Ras enhances signaling through the Ras/MAPK pathway and stimulates the deSUMOylation of MEK, SUMO of which initially inhibits its kinase activity. This leads to a phosphorylation cascade, in which Raf phosphorylates (P) MEK and phospho-MEK phosphorylates MAPK. Phospho-MAPK translocate to the nucleus and phosphorylates various transcription factors (TF) altering the transcriptional program of the cell.
Because of the role of Ras/MAPK signalling in directing the dorsal follicle cell fate, hypomorphic ras alleles lead to ventralized egg shells (i.e., fused dorsal appendages). Consistent with the idea that SUMO is required for Ras/MAPK signalling, reduced SUMO activity enhances this defect [24]. In addition, several proteins that modulate Ras signalling, including protein phosphatase 2A(PP2A) and two 14-3-3 family proteins, appear to be SUMO-conjugation targets [25]. Furthermore, RNAi knock down of SUMO in S2 cells reduces Ras/MAPK signalling in response to the RTK ligands insulin and Spitz as indicated by reduced levels of activated MEK and MAPK. PP2A and 14-3-3 family proteins are known to function via the Raf protein [26-30]. For example, PP2A in mammals is known to remove an inhibitory phosphate group from a serine residue in Ras, while 14-3-3 proteins bind to phosphoserine residues in Raf modulating its activity. Thus, these finding suggest that that SUMO influences Ras signalling upstream of MEK, perhaps at the level of Raf.
Studies in mammalian systems also demonstrate that SUMO interacts with the Ras/MAPK pathway. Of the three Ras isoforms in humans (HRas, KRas, and NRas), KRas is most frequently mutated in solid tumors [31]. KRas mutant colorectal cells (CRC) depend on Ubc9 for their clonogenic growth [32], as knocking down Ubc9 prevents CRC from establishing colonies. Furthermore, Ubc9 knockdown in CRC xenografts in mice induces apoptosis. SUMO-3 (one of four SUMO family proteins encoded in the human genome) appears to be conjugated to all three Ras isoforms in HEK293 cells at lysine 42 [33]. In addition, mutating lysine 42 to arginine reduces Ras/MAPK signalling in response to both wildtype and constitutively active Ras.
The above studies suggest that SUMO may have a conserved role in the activation of the Ras/MAPK pathway in Drosophila and mammals. However, other studies have demonstrated that conjugation of SUMO to MEK inhibits Ras/MAPK signalling [34]. SUMO appears to sequester MEK to the plasma membrane disrupting its ability to activate MAPK. Interestingly, activation of Ras abrogates MEK SUMOylation. Thus, SUMO appears to have complex roles, both positive and negative, in the regulation of Ras signalling (Figure 2).
Jun N-terminal Kinase
The Jun-N-terminal Kinase (JNK) pathway, another highly conserved MAPK signalling pathway, regulates multiple processes during Drosophila development, including dorsal closure in embryos, thorax closure in pupae, and stress induced apoptosis [35-37]. Intrinsic and external stimuli trigger the pathway by activating JNK Kinase (JNKKK), which then phosphorylates MAPK Kinase (MKK) for the subsequent phosphorylation and activation of JNK [38].
As mentioned above, the JNK pathway is known to upregulate apoptosis. SUMO was found to antagonize this process since SUMO knockdown by RNAi led to increased apoptosis in the wing disc [39]. However, when SUMO and JNK were knocked down at the same, increased apoptosis was not observed. Studies in human neuroblastoma SH-SY5Y cells also revealed that SUMO antagonizes JNK induced apoptosis [40].
Genetic and biochemical analyses reveal that SUMO modifies and regulates JNK activity via homeodomain-interacting protein kinase (HIPK) in Drosophila and humans [39,41]. SUMO modifies HIPK at a conserved lysine residue (Lys 25) [41]. Studies conducted in Drosophila and mammalian systems demonstrate that SUMO negatively regulates HIPK-mediated activation of apoptosis by retaining it in the nucleus [39,42]. Apparently, when cells are depleted of SUMO, HIPK enters the cytoplasm where it encounters and activates the JNK pathway leading to apoptosis.
Dpp signalling
Dpp signalling, which is required for many developmental pathways, including embryonic dorsoventral patterning and imaginal disc patterning, is also regulated by SUMO. In this case, SUMO appears to interfere with signalling. Dpp is a member of the BMP subfamily of TGFβ family ligands and signals through a heteromeric receptor consisting of a type I subunit (Saxophone or Thick veins) and a type II subunit (Punt) [43]. Both type I and type II subunits possess Ser/Thr kinase activity. After activation of the receptor by Dpp, the type II subunit phosphorylates the type I subunit, and then the type I subunit phosphorylates the Smad family transcription factor Mothers against Dpp (Mad). Phospho-Mad then interacts with the co-Smad Medea (Med) and activates downstream targets at the level of transcription [44].
A yeast two-hybrid screen uncovered an interaction between Med and Ubc9, and tissue culture experiments using S2 cells demonstrated Med SUMOylation [45]. Furthermore, overexpression of SUMO in the embryo inhibited the transcription of the Medea target genes Ance and ush, while expression of the two targets increased upon expression of a Med mutant containing a defective SUMO acceptor site. Fluorescent Recovery After Photobleaching (FRAP) studies showed that SUMOylation of Med occurs in the nucleus and allows for the shuttling of Med out of the nucleus, thus explaining how Med SUMOylation interferes with Dpp signalling.
Similarly, SUMO modifies the mammalian homologue of Med, Smad4 [46,47]. Lin and colleagues conducted a yeast two-hybrid screen, which revealed that SUMO-1 interacts with Smad4 and utilized site-directed mutagenesis to map the SUMO acceptor sites to lysines 113 and 159 [47]. Lee et al. also discovered that SUMO negatively regulates TGFb signalling through an assay in which four tandem Smad binding sites were fused to a luciferase reporter.
Mutagenesis of the SUMO-acceptor lysines in Smad4 enhanced TGFb induced expression of a reporter gene. Additional studies confirm that SUMO represses Smad4 transcriptional activity [48,49].
Polycomb group function
While the spatially regulated transcription factors (i.e., the products of the gap and pair rule genes) that initiate homeotic gene expression are only present in the early embryo, the spatially restricted patterns of homeotic gene expression are somehow maintained throughout embryonic and imaginal development. This cellular memory is thought to be provided by two groups of genes termed the Polycomb-group (PcG) and the Trithorax-group (TrxG), with the former being required for epigenetic stability of the repressed state, while the latter is required for epigenetic stability of the active state [50].
Many of the PcG proteins are members of one of three different complexes, the Pleiohomeotic Repressive Complex (PhoRC), Polycomb Repressive Complex 1 (PRC1), and Polycomb Repressive Complex 2 (PRC2) [51]. PhoRC, which contains Pleiohomeotic (Pho) and Scm-related gene containing four MBT domains (Sfmbt), binds to cis-regulatory elements in the homeotic gene complex termed Polycomb Response Elements (PREs), where they are thought to recruit PRC2. This complex contains Enhancer of zeste (E(z)) a SET family histone methyltransferase domain, which catalyses the trimethylation of lysine 27 on histone H3 (H3K27me3). H3K27me3 then serves as a docking site for PRC1. This complex ubiquitylates histone H2A and directs the compaction of chromatin, with this latter function serving to reduce the accessibility of associated genes to a TrxG-encoded chromatin remodeling complex that opens the chromatin allowing the transcriptional machinery to gain access to the DNA template. An additional PcG gene product that is essential for PcG function is Sex combs on midleg (Scm), which may be a peripheral component of PRC1.
Like its C. elegans homolog SOP-2 [52], Drosophila Scm is regulated by SUMO.
Knockdown of SUMO in S2 cells was found to increase association of Scm with a PRE in the homeotic gene complex and to result in the de-repression of the homeotic gene Ultrabithorax (Ubx). Conversely, knockdown of the SUMO deconjugating enzyme Ulp1 was found to decrease Scm association with the PRE. These findings are consistent with the idea that SUMO acts to negatively regulate Scm activity and, through Scm, to alleviate PcG-mediated repression. In support of this idea, mutagenesis of three consensus SUMO acceptor sites in Scm significantly reduced Scm SUMOylation and led to increased association of Scm with the PRE [53].
Consistent with the idea that SUMO negatively regulates Scm activity and therefore positively regulates Ubx expression, knockdown of SUMO in developing haltere discs results in an Ubx-like phenotype, i.e., a partial haltere-to-wing transformation.
The mechanism by which SUMO controls Scm and therefore polycomb group activity is unclear. Both Scm and the PRC1 component Polyhomeotic contain sterile alpha motif (SAM) domains, which are capable of mediating the formation of long protein filaments, and that may be required for chromatin compaction [54,55]. The functions of the Scm SAM domain are complex: it is required for recruitment of Ubc9 and thus SUMOylation, but it also appears to have an independent requirement in the recruitment of Scm to the PRE [53]. We speculate that Scm SUMOylation could modulate PcG function by modulating the role of the SAM domain in such processes as Scm recruitment, filament formation, and chromatin compaction.
In contrast to what is observed in Drosophila, in mammalian systems, SUMO has been shown to upregulate PcG activity. SUMO modifies the PcG protein Pc2 at lysine 492 and is necessary for its recruitment to PcG target genes [56,57]. In addition, mutation of SUMO-Specific Protease 2 (SENP2) enhances Pc2 recruitment to H3K27me3 and reduces the expression of PcG target genes, GATA4 and GATA 6, in mice [57].
Regulation of Innate immunity by SUMO via the Toll Pathway
Mammals have both innate and adaptive immune responses, and both work together to neutralize pathogens. The Drosophila innate immune system serves as a powerful model system for studying innate immunity since its effects are not masked by the adaptive immune response, which is absent in Drosophila. In both Drosophila and mammals, the innate immune response is regulated by signaling through both the Toll Like Receptor (TLR) and Immune Deficiency (IMD) pathways, which activate NF-?B-targeted antimicrobial peptide (AMPs) genes [58]. One Drosophila TLR, Toll, is activated by the ligand Spätzle [59,60]. This leads to the destabilization of the cytoplasmic I?B protein Cactus by the MyD88, Tube and Pelle Kinase complex [58]. The kinase complex phosphorylates Cactus to promote its degradation. IkB is an inhibitor of nuclear uptake of NF-?B family proteins, which includes three REL transcription factors, Dorsal, Dif, and Relish. Thus, Toll signaling leads to translocation of these proteins from the cytoplasm to the nucleus.
Several studies have demonstrated that SUMO upregulates TLR signaling induced transcription in both flies and mammals (Figure 3). In mammals, the SUMO protease, SUMO-specific Protease 6 (SENP6), down-regulates TLR activity [61]. Studies in Drosophila S2 cells demonstrate that SUMO is necessary for Dorsal to localize to the nucleus and activate the immune pathway [62]. Overexpression of SUMO in S2 cells, significantly increases the transactivation potential of Dorsal in a reporter assay. Furthermore, the AMPs, cecropin A1 (cecA1) and drosomycin (drs), which are dependent on rel family proteins such as Dorsal for transcriptional activation, also require SUMO for their expression in cultured cells. In addition, transcription of AMP genes in response to induction by the TLR agonist lipopolysaccharide is reduced in first instar larvae homozygous for sumo, ubc9, or dl mutations.
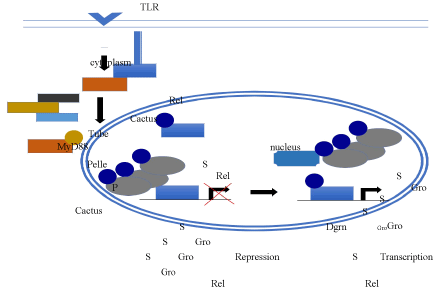
Figure 3. Regulation of Toll-Like Receptors by SUMO. Activation of the TLR promotes the Tube, MyD88, and Pelle complex to phosphorylate (P) Cactus and inhibit its ability to sequester the REL transcription factor in the cytoplasm. SUMOylation (S) of REL transcription factors promotes the translocation of the transcription factors from the cytoplasm and into the nucleus. At the same time SUMOylated Gro can interact with the transcription factors to inhibit transcription. However, Dgrn can interact with SUMOylated Groucho via a SUMO interaction motif to sequester the co-repressor from the transcription factor, thus allowing for transcriptional activation to occur.
Studies of Degringolade (Dgrn), a SUMO Targeted Ubiquitin Ligase (STUbL), also suggest a positive role for SUMO in the immune response [63,64]. Dgrn mutant flies are susceptible to fungal and bacterial infection since they fail to express AMPs. Furthermore, Dgrn alleviates inhibition by Cactus of Dorsal and Dif nuclear translocation upon stimulation of the Toll pathway.
Groucho, a transcriptional co-repressor, could provide a link between Dgrn and the innate immune response. Groucho, which functions, in part, by mediating the recruitment of Histone Deacetylase 1 (HDAC1) to its target genes, is required for repression by many of the transcriptional repressors that act throughout Drosophila development, including Dorsal [65,66-69]. Groucho is a SUMOylation target [25], and, in mammalian cells, SUMO appears to positively regulate Groucho function by helping to mediate the recruitment of HDAC1 through a SIM in
HDAC1 [70]. In contrast, work in Drosophila suggests that SUMO antagonizes Groucho-mediated repression. Dgrn binds Groucho in a SUMO dependent manner leading to the sequestration and therefore inactivation of Groucho. Thus, in the absence of SUMO, sequestration does not occur allowing for Groucho-mediated repression [64]. Since Groucho is a corepressor for Dorsal, this could inhibit activation of Dorsal-targeted AMP genes.
While the above studies suggest SUMO promotes Toll signalling in Drosophila innate immunity, SUMO also appears to negatively regulate the pathway via b-arrestin, which, in flies, is encoded by Kurz (Krz) [3,71]. Proteomic analysis shows that Krz interacts with Ulp1. In addition, Krz and Ulp1 knockdown larvae exhibit melanotic masses and increased lamellocyte differentiation in the haemolymph, phenotypes that are indicative of hyperactive Toll signalling [72-74]. Consistent with these observations, Krz and Ulp1 appear to work synergistically to deSUMOylate Dorsal. Furthermore, Dorsal and Dif are predominantly localized to the nucleus in Krz mutant larvae and this leads to increased Drs expression.
Concluding remarks
SUMO acts as a genetic switch that targets hundreds or thousands of proteins to regulate a wide variety of essential cellular and developmental processes. Illuminating its biological roles is as challenging as trying to arrive at a comprehensive understanding of the roles of other common protein modifications, such as phosphorylation, acetylation, and glycosylation [75]. Due to the pleiotropic functions of SUMO in development, global disruption of SUMOylation is not usually instructive. Therefore, approaches such as mapping and mutating individual SUMO acceptor sites, SUMO-substrate fusions, and tissue-specific overexpression or knockdown of SUMO pathway components must be utilized to dissect specific SUMO functions from one another.
Another challenge to understanding the many biological roles of SUMO is the so-called “SUMO enigma” [76]. In most cases, it appears that only a small fraction of any given SUMOylation target is conjugated to SUMO at any one time. Paradoxically, however, SUMOylation of proteins such as Scm, Groucho, HIPK, and Med often leads to near quantitative effects on the activity or subcellular localization of these proteins. While this enigma remains unresolved, two speculative non-mutually exclusive explanations are as follows. First, it possible that cyclic rounds of conjugation and deconjugation are required for progress through a pathway. Second, perhaps deconjugation leaves behind a protein that still retains the memory of being SUMOylated. For example, SUMOylation could be required to overcome a kinetic barrier to the formation of a protein complex that remains stable after deconjugation has occurred.
The ease with which the Drosophila genome can be manipulated has allowed us to overcome the challenges described above. Since pathways regulated by SUMO are highly conserved across the eukaryotic domain, studies of SUMOylation in Drosophila may provide insight into how SUMO leads to increased developmental complexity by diversifying protein function in mammals.
References
- Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, et al. (1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol 280: 275-286. [crossref]
- Smith M, Turki-Judeh W2, Courey AJ3 (2012) SUMOylation in Drosophila Development. Biomolecules 2: 331-349. [crossref]
- Anjum SG, Xu W, Nikkholgh N, Basu S, Nie Y, et al. (2013) Regulation of Toll signaling and inflammation by β-arrestin and the SUMO protease Ulp1. Genetics 195: 1307-1317. [crossref]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109-120. [crossref]
- Schmidt D, Müller S (2002) Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci U S A 99: 2872-2877. [crossref]
- Agrawal N, Banerjee R (2008) Human polycomb 2 protein is a SUMO E3 ligase and alleviates substrate-induced inhibition of cystathionine beta-synthase sumoylation. PLoS One 3: e4032.
- Rodriguez MS, Dargemont C, Hay RT (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 276: 12654-12659. [crossref]
- Smith M, Bhaskar V, Fernandez J, Courey AJ (2004) Drosophila Ulp1, a nuclear pore-associated SUMO protease, prevents accumulation of cytoplasmic SUMO conjugates. J Biol Chem 279: 43805-43814. [crossref]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A 101: 14373-14378. [crossref]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I (2006) Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281: 16117-16127. [crossref]
- Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, et al. (2005) Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435: 979-982. [crossref]
- Kerscher O (2007) SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep 8: 550-555. [crossref]
- Reeves GT, Stathopoulos A (2009) Graded dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb Perspect Biol 1: a000836. [crossref]
- Pawson T, Gish GD (1992) SH2 and SH3 domains: from structure to function. Cell 71: 359-362. [crossref]
- Simon MA, Bowtell DDL, Dodson GS, Laverty TR, Rubin GM (1991) Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 67: 701-716.
- Simon MA, Dodson GS, Rubin GM (1993) An SH3-SH2-SH3 protein is required for p21Ras1 activation and binds to sevenless and Sos proteins in vitro. Cell 73: 169-177. [crossref]
- Bonfini L, Karlovich CA, Dasgupta C, Banerjee U (1992) The Son of sevenless gene product: a putative activator of Ras. Science 255: 603-606. [crossref]
- Leevers SJ, Paterson HF, Marshall CJ (1994) Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature 369: 411-414. [crossref]
- Wellbrock C, Karasarides M, Marais R (2004) The RAF proteins take centre stage. Nat Rev Mol Cell Biol 5: 875-885. [crossref]
- McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, et al. (2007) Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta 1773: 1263-1284. [crossref]
- Hsu JC, Perrimon N (1994) A temperature-sensitive MEK mutation demonstrates the conservation of the signaling pathways activated by receptor tyrosine kinases. Genes & Development 8: 2176-2187.
- Schnorr JD, Berg CA (1996) Differential activity of Ras1 during patterning of the Drosophila dorsoventral axis. Genetics 144: 1545-1557. [crossref]
- Brand AH, Perrimon N (1994) Raf acts downstream of the EGF receptor to determine dorsoventral polarity during Drosophila oogenesis. Genes Dev 8: 629-639. [crossref]
- Schnorr JD, Holdcraft R, Chevalier B, Berg CA (2001) Ras1 interacts with multiple new signaling and cytoskeletal loci in Drosophila eggshell patterning and morphogenesis. Genetics 159: 609-622. [crossref]
- Nie M, Xie Y, Loo JA, Courey AJ (2009) Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS One 4: e5905. [crossref]
- Abraham D, Podar K, Pacher M, Kubicek M, Welzel N, et al. (2000) Raf-1-associated protein phosphatase 2A as a positive regulator of kinase activation. J Biol Chem 275: 22300-22304. [crossref]
- Light Y, Paterson H, Marais R (2002) 14-3-3 antagonizes Ras-mediated Raf-1 recruitment to the plasma membrane to maintain signaling fidelity. Mol Cell Biol 22: 4984-4996. [crossref]
- Rommel C, Radziwill2021 Copyright OAT. All rights reserv, Heinicke T, et al. (1996) Activated Ras displaces 14-3-3 protein from the amino terminus of c-Raf-1. Oncogene 12: 609-619. [crossref]
- Rommel C, Radziwill G, Moelling K, Hafen E (1997) Negative regulation of Raf activity by bindizg of 14-3-3 to the amino terminus of Raf in vivo. Mech Dev 64: 95-104. [crossref]
- Wassarman DA, Solomon NM, Chang HC, Karim FD, Therrien M, et al. (1996) Protein phosphatase 2A positively and negatively regulates Ras1-mediated photoreceptor development in Drosophila. Genes & Development 10: 272-278.
- Zhang H, Luo J (2016) SUMO wrestling with Ras. Small GTPases 7: 39-46. [crossref]
- Yu B, Swatkoski S, Holly A3, Lee LC1, Giroux V1, et al. (2015) Oncogenesis driven by the Ras/Raf pathway requires the SUMO E2 ligase Ubc9. Proc Natl Acad Sci U S A 112: E1724-1733. [crossref]
- Choi BH, Chen C2, Philips M3, Dai W1,4 (2017) RAS GTPases are modified by SUMOylation. Oncotarget 9: 4440-4450. [crossref]
- Kubota Y, O'Grady P, Saito H, Takekawa M (2011) Oncogenic Ras abrogates MEK SUMOylation that suppresses the ERK pathway and cell transformation. Nat Cell Biol 13: 282-291. [crossref]
- Etter PD, Narayanan R, Navratilova Z, Patel C, Bohmann D, et al. (2005) Synaptic and genomic responses to JNK and AP-1 signaling in Drosophila neurons. BMC Neurosci 6: 39. [crossref]
- Luo X, Puig O, Hyun J, Bohmann D, Jasper H (2007) Foxo and Fos regulate the decision between cell death and survival in response to UV irradiation. EMBO J 26: 380-390. [crossref]
- Igaki T (2009) Correcting developmental errors by apoptosis: lessons from Drosophila JNK signaling. Apoptosis: an international journal on programmed cell death 14: 1021-1028.
- Biteau B, Karpac J, Hwangbo D, Jasper H (2011) Regulation of Drosophila lifespan by JNK signaling. Exp Gerontol 46: 349-354. [crossref]
- Huang H, Du G, Chen H, Liang X, Li C, Zhu N, et al. (2011) Drosophila Smt3 negatively regulates JNK signaling through sequestering Hipk in the nucleus. Development (Cambridge, England) 138: 2477-85.
- Feligioni M, Brambilla E, Camassa A, Sclip A, Arnaboldi A, Morelli F, et al. (2011) Crosstalk between JNK and SUMO signaling pathways: deSUMOylation is protective against H2O2-induced cell injury. PLoS One 6: e28185.
- Hofmann TG, Jaffray E, Stollberg N, Hay RT, Will H (2005) Regulation of homeodomain-interacting protein kinase 2 (HIPK2) effector function through dynamic small ubiquitin-related modifier-1 (SUMO-1) modification. J Biol Chem 280: 29224-29232. [crossref]
- de la Vega L, Fröbius K, Moreno R, Calzado MA, Geng H, et al. (2011) Control of nuclear HIPK2 localization and function by a SUMO interaction motif. Biochim Biophys Acta 1813: 283-297. [crossref]
- Shimmi O, Umulis D, Othmer H, O'Connor MB (2005) Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell 120: 873-886. [crossref]
- Affolter M, Marty T, Vigano MA, Jaźwińska A (2001) Nuclear interpretation of Dpp signaling in Drosophila. EMBO J 20: 3298-3305. [crossref]
- Miles WO, Jaffray E, Campbell SG, Takeda S, Bayston LJ, Basu SP, et al (2008) Medea SUMOylation restricts the signaling range of the Dpp morphogen in the Drosophila embryo. Genes & Development 22: 2578-2590.
- Lee PS, Chang C, Liu D, Derynck R (2003) Sumoylation of Smad4, the common Smad mediator of transforming growth factor-beta family signaling. J Biol Chem 278: 27853-27863. [crossref]
- Lin X, Liang M, Liang Y-Y, Brunicardi FC, Melchior F, Feng X-H (2003) Activation of Transforming Growth Factor-ß Signaling by SUMO-1 Modification of Tumor Suppressor Smad4/DPC4. J Biological Chemistry 278: 18714-18719.
- Long J, Wang G, He D, Liu F (2004) Repression of Smad4 transcriptional activity by SUMO modification. Biochem J 379: 23-29. [crossref]
- Yukita A, Hosoya A, Ito Y, Katagiri T, Asashima M, Nakamura H (2012) Ubc9 negatively regulates BMP-mediated osteoblastic differentiation in cultured cells. Bone 50: 1092-1099.
- Schuettengruber B, Bourbon HM2, Di Croce L3, Cavalli G4 (2017) Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171: 34-57. [crossref]
- Schwartz YB, Pirrotta V (2013) A new world of Polycombs: unexpected partnerships and emerging functions. Nat Rev Genet 14: 853-864. [crossref]
- Zhang H, Smolen GA, Palmer R, Christoforou A, van den Heuvel S, et al. (2004) SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans Polycomb group protein SOP-2. Nat Genet 36: 507-511. [crossref]
- Smith M, Mallin DR, Simon JA, Courey AJ (2011) Small ubiquitin-like modifier (SUMO) conjugation impedes transcriptional silencing by the polycomb group repressor Sex Comb on Midleg. J Biol Chem 286: 11391-11400. [crossref]
- Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, et al. (2016) Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529: 418-422.
- Peterson AJ, Mallin DR, Francis NJ, Ketel CS, Stamm J, et al. (2004) Requirement for sex comb on midleg protein interactions in Drosophila polycomb group repression. Genetics 167: 1225-1239. [crossref]
- Roscic A, Möller A, Calzado MA, Renner F, Wimmer VC, et al. (2006) Phosphorylation-Dependent Control of Pc2 SUMO E3 Ligase Activity by Its Substrate Protein HIPK2. Molecular cell 24: 77-89.
- Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, et al. (2010) SUMO-Specific Protease 2 Is Essential for Suppression of Polycomb Group Protein-Mediated Gene Silencing during Embryonic Development. Molecular cell 38: 191-201.
- Govind S (2008) Innate immunity in Drosophila: Pathogens and pathways. Insect Sci 15: 29-43. [crossref]
- Morisato D, Anderson KV (1994) The spätzle gene encodes a component of the extracellular signaling pathway establishing the dorsal-ventral pattern of the Drosophila embryo. Cell 76: 677-688. [crossref]
- Schneider DS, Jin Y, Morisato D, Anderson KV (1994) A processed form of the Spatzle protein defines dorsal-ventral polarity in the Drosophila embryo. Development (Cambridge, England) 120: 1243-50.
- Liu X, Chen W, Wang Q, Li L, Wang C (2013) Negative regulation of TLR inflammatory signaling by the SUMO-deconjugating enzyme SENP6. PLoS Pathog 9: e1003480. [crossref]
- Bhaskar V, Smith M, Courey AJ (2002) Conjugation of Smt3 to dorsal may potentiate the Drosophila immune response. Mol Cell Biol 22: 492-504. [crossref]
- Koltun B, Shackelford E, Bonnay F, Matt N, Reichhart JM, Orian A (2017) The SUMO-targeted ubiquitin ligase, Dgrn, is essential for Drosophila innate immunity. Inter J developmental biology 61: 319-27.
- Abed M, Barry KC, Kenyagin D, Koltun B, Phippen TM, et al. (2011) Degringolade, a SUMO-targeted ubiquitin ligase, inhibits Hairy/Groucho-mediated repression. The EMBO journal 30:1289-301.
- Turki-Judeh W, Courey AJ (2012) Groucho: a corepressor with instructive roles in development. Curr Top Dev Biol 98: 65-96. [crossref]
- Dubnicoff T, Valentine SA, Chen G, Shi T, Lengyel JA, et al. (1997) Conversion of dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev 11: 2952-2957. [crossref]
- Hasson P, Müller B, Basler K, Paroush Z (2001) Brinker requires two corepressors for maximal and versatile repression in Dpp signalling. EMBO J 20: 5725-5736. [crossref]
- Jiménez G, Paroush Z, Ish-Horowicz D (1997) Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev 11: 3072-3082. [crossref]
- Paroush Z, Finley RL, Jr., Kidd T, Wainwright SM, Ingham PW, et al. (1994) Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79: 805-815.
- Ahn JW, Lee YA, Ahn JH, Choi CY (2009) Covalent conjugation of Groucho with SUMO-1 modulates its corepressor activity. Biochem Biophys Res Commun 379: 160-165. [crossref]
- Tipping M, Kim Y, Kyriakakis P, Tong M, Shvartsman SY, et al. (2010) β-arrestin Kurtz inhibits MAPK and Toll signalling in Drosophila development. EMBO J 29: 3222-3235. [crossref]
- Gerttula S, Jin YS, Anderson KV (1988) Zygotic expression and activity of the Drosophila Toll gene, a gene required maternally for embryonic dorsal-ventral pattern formation. Genetics 119: 123-133. [crossref]
- Lemaitre B, Meister M, Govind S, Georgel P, Steward R, et al. (1995) Functional analysis and regulation of nuclear import of dorsal during the immune response in Drosophila. EMBO J 14: 536-545. [crossref]
- Roth S, Hiromi Y, Godt D, Nusslein-Volhard C (1991) Cactus, a maternal gene required for proper formation of the dorsoventral morphogen gradient in Drosophila embryos. Development (Cambridge, England) 112: 371-88.
- Lomelí H, Vázquez M (2011) Emerging roles of the SUMO pathway in development. Cell Mol Life Sci 68: 4045-4064. [crossref]
- Hay RT1 (2005) SUMO: a history of modification. Mol Cell 18: 1-12. [crossref]



