Alternative therapy with medical devices using high-voltage electric potential (HELP) to generate an electric field (EF) is common in Japan. However, the mechanisms underlying potential health benefits of this therapy remain unclear. Therefore, we investigated the effect of HELP exposure (9 kV /electrode + 9 kV/electrode, 30 min) on N-acyl serines (N-acyl SERs) using selected reaction monitoring (SRM) analysis in plasma samples obtained from healthy human subjects before and after a single treatment session. N-18:1 SER, and N-16:0 SER were significantly upregulated following HELP exposure. Under these conditions, HELP exposure did not exert on levels of N-18:0 SER, N-18:2 SER, N-20:4 SER, and N-22:6 SER. Because N-18:1 ethanolamine (N-18:1 EA) is known to activate, transient receptor potential vanilloid 1 (TRPV1) and peroxisome proliferator-activated receptor-alpha (PPAR-α), we further examined an in silico docking simulation with TRPV1 and PPAR-α. The binding energies with TRPV1 were -6.359 and -6.227 kcal/mol for N-18:1 SER and N-18:1 EA, respectively. The binding energies with PPAR-α were -7.366 and -6.956 kcal/mol for N-18:1 SER and N-18:1 EA, respectively. In human HepG2 cells, N-18:1 SER enhanced fatty acid-binding protein 1 (FABP1) mRNA expression. N-18:1 SER-induced FABP1 mRNA expression was sensitive to the PPAR-α antagonist GW6471. Our findings provide new insights into the molecular mechanisms of the health benefits of EF therapy.
N-oleoyl serine, N-palmitoyl serine, N-oleoyl ethanolamine, TRPV1, PPAR-a
Abbreviations
ACOX1: peroxisomal acyl-coenzyme A oxidase 1; DSPS: delayed sleep phase syndrome; EC50: half maximal effective concentration; EF: electric field; FA: fatty acid; FABP1: fatty acid-binding protein 1; HELP: high-voltage electric potential; HODE: hydroxyoctadecadienoic acid; IL: interleukin; MSL: multiple benign symmetric lipomatosis; N-18:2 EA: N-linoleoyl ethanolamine; N-18:1 EA: N-oleoyl ethanolamine; N-16:0 EA: N-palmitoyl ethanolamine; N-18:0 EA: N-stearoyl ethanolamine; NF-κB: nuclear factor-kappa B; NLRP3: nod-like receptor protein 3; N-20:4 SER: N-arachidonyl serine; N-22:6 SER: N-docosahexaenoyl serine; N-18:2 SER: N-linoleoyl serine; N-18:1 SER: N-oleoyl serine; N-16:0 SER: N-palmitoyl serine; N-18:0 SER: N-stearoyl serine; PLAAT: phospholipase A/acyltransferase; PLA2: phospholipase A2; PPAR-α: peroxisome proliferator-activated receptor-alpha; qRT-PCR: quantitative real-time polymerase chain reaction; SRM: selected reaction monitoring; TLR4: toll-like receptor 4; TRPV1: transient receptor potential vanilloid 1.
High-voltage electric field (EF) therapy may be an effective treatment for shoulder stiffness, headache, insomnia, and chronic constipation [1-19]. Although EF therapy was discovered about 90 years ago, the molecular mechanisms associated with its health benefits remain elusive. A therapeutic device that exposes the human body to high-voltage electric potential (HELP) was approved by the Ministry of Health, Labour and Welfare in Japan [1-19]. A review of the literature suggests that HELP exposure may be an alternative therapy for several conditions. Our previous attempts at finding HELP exposure-induced plasma biomarkers, using liquid chromatography (LC)-time-of-flight mass spectrometry, led to the detection of lipids such as palmitic acid, palmitoleic acid, oleic acid, linoleic acid, linolenic acid, cis-8,11,14-eicosatrienoic acid (DGLA), cis-5,8,11,14,17-eicosapentaenoic acid, and cis-4,7,10,13,16,19-docosahexaenoic acid [12]. Recent methods for quantifying lipid mediators have taken advantage of selected reaction monitoring (SRM) analysis [17,18]. Using SRM, we showed enhanced levels of HELP exposure-induced lipids, including lysophosphatidylcholine-22:4, lysophosphatidylethanolamine-20:4, and lysophosphatidylethanolamine-22:6 in the plasma of healthy individuals [17,18]. Therefore, we hypothesized that changes in plasma lyso-form of phosphatidylethanolamine levels after EF exposure may be linked to changes in N-acyl serines (N-acyl SERs). The stimulatory effects of N-acyl SERs, such as N-18:1 SER and N-16:0 SER on cell number in osteoblastic MC3T3 E1 cells, reported recently [20], prompted an investigation into detection of plasma N-acyl SERs using SRM in healthy subjects subjected to a single HELP stimulation. In this study, we demonstrated that N-18:1 SER and N-16:0 SER were upregulated by HELP exposure (9 kV/electrode + 9 kV/electrode, 30 min). In addition, we conducted an in silico docking simulation to explore the interactions between N-18:1 SER or N-16:0 SER and the active site of TRPV1, or PPAR-α
EF exposure
As described previously [1-19], EF exposure comprised of a system that was equipped with a transformer, a seat, and two insulator-covered electrodes. One electrode was placed on the floor plate, near the subject’s feet, while the other was placed on the head. An EF generated by the HELP apparatus (Healthtron PRO-18T; Hakuju Institute for Health Science Co., Ltd., Tokyo, Japan) was uniformly created by transforming a 50-Hz alternating current at 9 kV/electrode + 9 kV/electrode. The Japanese government confirmed this system was safe for human use in 1963.
Subjects
Fifty-seven healthy adults (23 males and 34 females; mean age, 46.6 ± 0.9 years; body mass index [BMI], 22.1 ± 0.6 kg/m2) volunteered for experiment 1 (exposure condition: 9 kV/electrode + 9 kV/electrode, 30 min). Thirty-two healthy adults (11 males and 21 females; mean age, 46.4 ± 1.3 years; BMI, 22.5 ± 0.9 kg/m2) volunteered for experiment 2 (exposure condition: 9 kV/electrode + 9 kV/electrode, 30 min). All participants signed an informed consent form after receiving verbal and written information about the study. All experiments were performed in the morning. All experiments were conducted in accordance with the Declaration of Helsinki. The study protocol was approved by the human ethics committee of Hakuju Institute for Health Science Co., Ltd. (Tokyo, Japan).
Plasma preparation
Blood samples were collected using vacutainer tubes coated with ethylene diamine tetra acetic acid (VP-NA070K; Terumo Corporation, Tokyo, Japan) and immediately centrifuged at 800 x g for 5 min to separate the plasma from other cellular materials. The plasma was subsequently transferred to a fresh Eppendorf tube and stored at -80℃ until processing.
Solid-phase extraction of N-acyl amides from human plasma
Solid-phase extraction of N-acyl SERs and N-acyl ethanolamines (N-acyl EAs) were performed, as previously reported, with minor modifications [21]. Briefly, 5 pmol N-arachidonyl-L-serine-d8 (Cayman Chemicals, Ann Arbor, MI, USA) and 25 pmol N-arachidonyl ethanolamide-d8 (Cayman Chemicals, Ann Arbor, MI, USA) were added to plasma (250 µL) as an internal standard. Protein was precipitated by adding 1 ml of acetonitrile on ice for 15 min, followed by centrifuging at 5,000 rpm for 5 min at 4 °C. The supernatant was diluted with 0.005N HCl in water. Oasis HLB 60 mg Vac-RC columns (Waters, Milford, MA, USA) were preconditioned with methanol and 0.005N HCl in water via a vacuum manifold (Waters, Milford, MA, USA). The diluted extract was applied to the column and washed with 3 ml of 0.005N HCl in water, followed by 20% (v/v) acetonitrile. Finally the N-acyl amides were eluted from the column using 1 ml of acetonitrile and the fraction was dried under a constant stream of N2 gas, reconstituted in 40 µL of 70% (v/v) methanol, and transferred to sample vials.
LC-MS/MS analysis
A 20 µL aliquot of the lipid sample was injected and the lipids were separated on a Waters X-Bridge C18 (3.5 µm, 150 mm X 1.0 mm i.d., Waters Corporation, Milford, MA, USA) at 30℃ using a gradient solvent system as follows: mobile phase A [acetonitrile/methanol (4:1, v/v)] / mobile phase B [LC-MS grade distilled water supplemented with 0.1% acetic acid] ratios of 50%/50% (0-5 min), 50%/50% (5 min), 100%/0% (20-25 min), 50%/50% (25-30 min). Flow rate was 80 µL/min. N-acyl amides were measured by SRM in negative mode for N-acyl SERs, in positive ion mode for N-acyl EAs with a liquid chromatograph mass spectrometer LCMS-8040 (Shimadzu, Japan). SRM m/z transitions were: N-16:0 SER = 342.3/74.1; N-18:0 SER = 370.3/74.1; N-18:1 SER = 368.3/74.1; N-18:2 SER = 366.3/74.1; N-20:4 SER = 390.3/74.1; N-22:6 SER = 414.3/74.1; N-16:0 EA = 300.3/62.2; N-18:0 EA = 328.3/62.2; N-18:1 EA = 326.3/62.2; N-18:2 EA = 324.3/62.2.
Homology modeling and docking simulation
We used Q8NER1.fasta registered in UniProt to obtain the sequence of human TRPV1 (hTRPV1). The three-dimensional structure (3J5R; Protein Data Bank Japan) was used as a template structure. The best docking score was chosen from the docking calculation between capsaicin (TRPV1 agonist) and a homology model. The docking study of N-acyl SERs binding to the target protein of the hTRPV1 model structure was performed using the AutoDock Vina docking software (Dr. Oleg Trott, The Scripps Research Institute, CA, USA) [22]. The docking experiment was performed five times and yielded 100 candidate conformations.
Docking study on the nuclear PPAR- α
The X-ray crystal structure of PPAR-α complex with GW735 (2P54; Protein Data Bank Japan) was used for the molecular docking [23]. The docking study of N-acyl SERs binding to the target protein of the hPPAR- α model structure was performed using AutoDock Vina docking software (Dr. Oleg Trott, The Scripps Research Institute, CA, USA) [23]. The docking experiment was performed five times and yielded 100 candidate conformations.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Human hepatocellular carcinoma HepG2 cells were obtained from ATCC (HB-8065, Rockville, MD, USA). The cells were cultured in RPMI-1640 (Nakalai Tesque, Kyoto, Japan) containing 10% fetal calf serum (Sigma-Aldrich, St, Louis, MO, USA), 100 U/mL penicillin and 100 µg/mL streptomycin (Nacalai Tesque, Kyoto, Japan) in a humidified 5% CO2 atmosphere at 37oC. HepG2 cells (30,000 cells/well) were subcultured and seeded on 96-well plates (Sumitomo bakelite, Tokyo, Japan). After 24h incubation, the cells were treated with either N-oleoyl-L-serine (Cayman Chemical, Ann Arbor, MI, USA), or GW6471 (Sigma-Aldrich, St. Louis, MO, USA) or both for 24h. The cells treated with vehicle (0.5 % ethanol) were the used as the control. qRT-PCR was performed as previously described [12,15]. Total RNA was isolated using a FastLane cell cDNA kit (Qiagen, Valencia, CA, USA) and was reversely transcribed to complementary DNA (cDNA) using QuantiTect Reverse Transcription kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. qRT-PCR was carried out in a LightCyler 96 System (Roche Applied Science, Mannheim, Germany) using SYBR Green Premix Ex Taq II (Takara Bio Inc., Otsu, Japan). The gene expression level was calculated by the delta-delta-Ct method. Data were presented as relative expression units after normalization to the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene. The sequences (5’ to 3’) of primer sets were FABP1-F, ATG AGT TTC TCC GGC AAG TAC C; FABP1-R, CTC TTC CGG CAG ACC GAT TG; ACOX1-F, GTA TGG AAT CAG TCA GAA CGC; ACOX1-R, CTT GTA AGA TTC GTG GAC CTC; and GAPDH-F, CAT CCC TGC CTC TAC TGG CGC TGC C; GAPDH-R, CCA GGA TGC CCT TGA GGG GGC CCT C;
Statistical analysis
Data were analyzed using Welch’s t-test. A probability (p) value < 0.05 was considered statistically significant.
Effect of HELP exposure on N-acyl SERs in the plasma of healthy individuals
We analyzed the effect of HELP exposure (9 kV/electrode + 9 kV/electrode) for 30 min on N-acyl SERs (Figure 1). Plasma levels of N-16:0 SER and N-18:1 SER were significantly upregulated at the time-0 after HELP exposure compared with levels before exposure (N-16:0 SER: 1.42-fold, p=0.0305; N-18:1 SER: 1.49-fold, p=0.0315). Under these conditions, HELP exposure did not affect the levels of N-18:0 SER, N-18:2 SER, N-20:4 SER, and N-22:6 SER (Figure 1).

Figure 1. Effect of HELP exposure (9 kV/electrode + 9 kV/electrode, 30 min) on N-acyl SERs in the plasma of healthy individuals. (a) Typical N-18:1 SER peak in the plasma of healthy humans. N-18:1 SER was detected by SRM analysis. (b) Structure of N-18:1 SER. (c) Typical N-16:0 SER peak detected by SRM analysis. (d) Structure of N-16:0 SER. (e) N-Acyl SERs in plasma before and after exposure. Data are expressed as mean ± SE (n=57 in N-16:0 SER; n=43 in N-18:0 SER; n=57 in N-18:1 SER; n=52 in N-18:2 SER; n=24 in N-20:4 SER; n=40 in N-22:6 SER). N-16:0 SER is expressed as mean ± SE in the inset. * p < 0.05 compared with before exposure
Effect of HELP exposure on N-acyl EAs in the plasma of healthy individuals
Because N-acyl SERs are structurally similar to N-acyl EAs, we next analyzed the effect of HELP exposure (9 kV/electrode + 9 kV/electrode) for 30 min on N-acyl EAs (Figure 2). Plasma levels of N-16:0 EA, N-18:1 EA, and N-18:2 EA were significantly upregulated at the 30-min time point after HELP exposure compared with those observed before the exposure (N-16:0 EA: 1.24-fold, p=0.0300; N-18:1 EA: 1.27-fold, p=0.0285; N-18:2 EA: 1.34-fold, p=0.0170). Under these conditions, HELP exposure did not affect the level of N-18:0 EA (Figure 2).
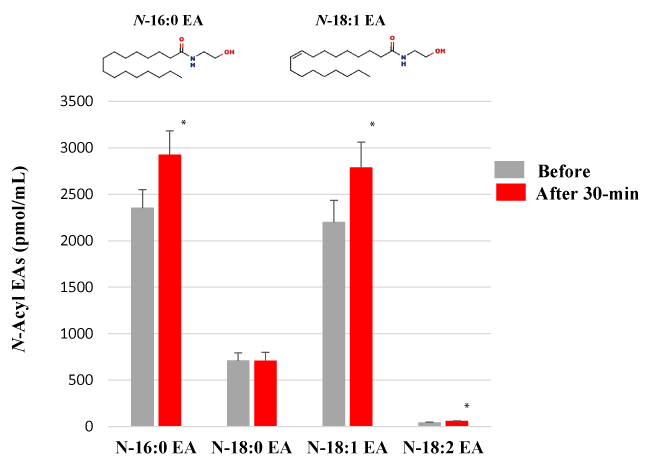
Figure 2. Effect of HELP exposure (9 kV/electrode + 9 kV/electrode, 30 min) on N-acyl EAs in the plasma of healthy individuals. N-Acyl EAs in plasma. Data are expressed as mean ± SE (n=32 in N-16:0 EA of before and after 30-min; n=32 in N-18:0 EA of before and after 30-min; n=32 in N-18:1 EA of before and after 30-min; n=27 in N-18:2 EA of before and after 30-min). * p < 0.05 compared with before exposure
Docking simulation of N-18:1 SER, N-16:0 SER, N-18:1 EA, N-16:0 EA, or N-18:2 EA with a homology model of TRPV1
N-18:1 EA and N-18:2 EA evoke an increase in intracellular calcium in HEK293 cells expressing hTRPV1 [24]. Therefore, we hypothesized that increased plasma N-18:1 SER and N-16:0 SER levels after HELP exposure may be linked to its activation as an endogenous agonist of TRPV1. We examined the in silico docking of N-18:1 SER, N-16:0 SER, or a well-known TRPV1 agonist, capsaicin, in the active site of TRPV1 using AutoDock Vina software [25-27]. We set the number of outputs poses to 20, with a total of 100 candidate conformations. Capsaicin showed a strong interaction energy of -7.915 kcal/mol (Table 1). Capsaicin formed hydrogen bonds with Thr-550, Ser-512, and Glu-570 (Figure 3a, Table 1). Under these conditions, N-18:1 SER showed good binding energy of -6.359 kcal/mol (Table 1). N-18:1 SER formed hydrogen bonds with Thr-550, and Ala-666 (Table 1, Figure 3b). A similar docking score was obtained using 9-HODE instead of N-18:1 SER (Table 1). 9-HODE, an endogenous agonist of TRPV1, formed hydrogen bonds with Thr-550, and Ala-666 (Table 1, Figure 3c). In addition, N-16:0 SER showed good binding energy of -6.077 kcal/mol (Table 1). N-16:0 SER formed hydrogen bonds with Thr-550, and Ala-666 (Table 1, Figure 3d). Subsequently, we examined the in silico docking of N-acyl EAs to the active site of TRPV1 to determine specificity. The binding energies were -6.421, -6.227, and -5.767 kcal/mol for N-18:2 EA, N-18:1 EA, and N-16:0 EA, respectively (Table 1).
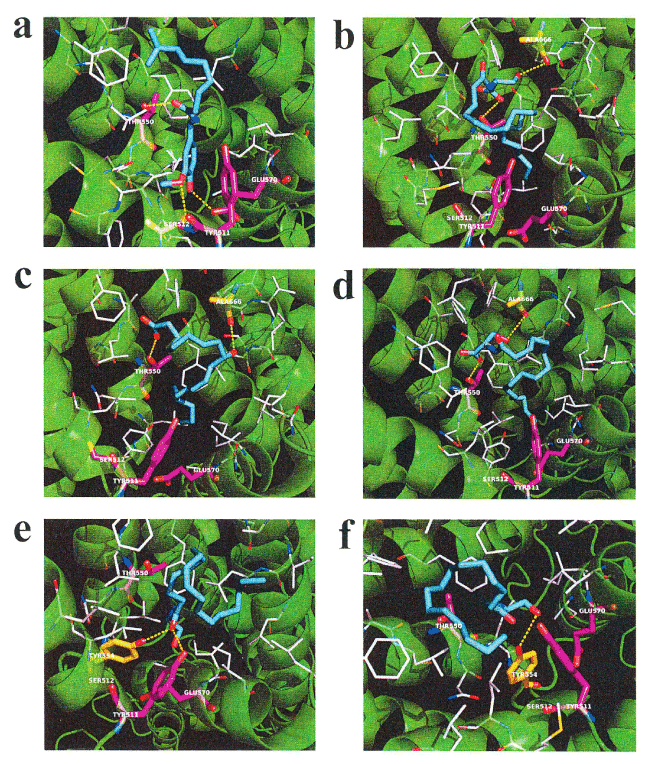
Figure 3. In silico molecular docking of N-18:1 SER, N-16:0 SER, N-18:1 EA, N-16:0 EA, 9-HODE, or capsaicin with TRPV1. (a) Binding mode of capsaicin in homology modeling of TRPV1; capsaicin (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The carboxyl of the capsaicin amide moiety forms a hydrogen bond with the hydroxyl group in the Thr-550 side chain. The methoxy group of the capsaicin vanilloid moiety forms a hydrogen bond with the hydroxyl group in the side chain of Ser-512. The hydroxyl group of the capsaicin vanilloid moiety forms a hydrogen bond with the hydroxyl group in the side chain of Glu-570. (b) Binding mode of N-18:1 SER in homology modeling of TRPV1. N-18:1 SER (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The carboxyl of the N-18:1 SER amide moiety forms a hydrogen bond with the hydroxyl group in the side chain of Thr-550. The hydroxyl group of the N-18:1 SER serine moiety forms a hydrogen bond with the carbonyl group in the main chain of Ala-666. (c) Binding mode of 9-HODE in homology modeling of TRPV1; 9-HODE (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The oxygen atom in the carboxyl group of 9-HODE forms a hydrogen bond with the hydroxyl group of Thr-550. The hydroxyl group of 9-HODE forms a hydrogen bond with the carbonyl in the main chain of Ala666. (d) Binding mode of N-16:0 SER in homology modeling of TRPV1; N-16:0 SER (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The carboxyl of the N-16:0 SER amide moiety forms a hydrogen bond with the hydroxyl group in the Thr-550 side chain. The hydroxyl group of the N-16:0 SER serine moiety forms a hydrogen bond with the carbonyl in the main chain of Ala-666. (e) Binding mode of N-18:1 EA in homology modeling of TRPV1; N-18:1 EA (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The carboxyl group in the N-18:1 EA amide forms a hydrogen bond with the Tyr-511 hydroxyl group. The carboxyl group in the N-18:1 EA amide forms a hydrogen bond with the Tyr-554 hydroxyl group. (f) Binding mode of N-16:0 EA in homology modeling of TRPV1; N-16:0 EA (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The hydroxyl group of N-16:0 EA forms a hydrogen bond with the Tyr-554 hydroxyl group
Table 1. Docking score and key interacting residues of TRPV1
Ligand |
Docking score |
Interactive residues |
|
(kcal/mol) |
|
TRPV1 agonist |
Capsaicin |
-7.915 |
Ser-512, Thr-550, and Glu-570 |
Endogenous TRPV1 agonist |
9-HODE |
-6.496 |
Thr-550, and Ala-666 |
N-Acyl serine |
|
|
N-18:1 SER |
-6.359 |
Thr-550, and Ala-666 |
N-16:0 SER |
-6.077 |
Thr-550, and Ala-666 |
N-Acyl ethanolamine |
N-18:1 EA |
-6.227 |
Tyr-511, and Tyr-554 |
N-16:0 EA |
-5.767 |
Tyr-554 |
N-18:2 EA |
-6.421 |
Glu-570 |
Docking simulation of N-18:1 SER, N-16:0 SER, N-18:1 EA, N-16:0 EA, or N-18:2 EA with a model of PPAR- α
N-18: EA is known to induce lipolysis as a putative endogenous activator of PPAR-α [13, 28-29]. We examined the in silico docking of N-18:1 SER, N-16:0 SER, or a well-known PPAR- α agonist, bezafibrate, in the active site of PPAR- α using AutoDock Vina software [30]. We set the number of outputs poses to 20, with a total of 100 candidate conformations. Bezafibrate showed a strong interaction energy of —9.323 kcal/mol (Table 2). Bezafibrate formed hydrogen bonds with Ser-280, Tyr-314, His-440, and Tyr-464 (Figure 4a, Table 2). Under these conditions, N-18:1 SER showed good binding energy of —7.366 kcal/mol (Table 2). N-18:1 SER formed hydrogen bonds with Ser-280, Tyr-314, and His-440 (Figure 4b, Table 2). In addition, N-16:0 SER showed good binding energy of -7.161 kcal/mol (Table 2). N-16:0 SER formed hydrogen bonds with Ser-280, Tyr-314, and Tyr-464 (Figure 4c, Table 2). A similar docking score was obtained using N-18:1 EA instead of N-16:0 SER (Table 2). N-18:1 EA formed hydrogen bonds with Ser-280, Tyr-314, and His-440 (Figure 4d, Table 2). Subsequently, we examined the in silico docking of N-16:0 EA and N-18:2 EA to the active site of PPAR-α to determine specificity. The binding energies were -6.570 and -6.979, respectively (Table 2).
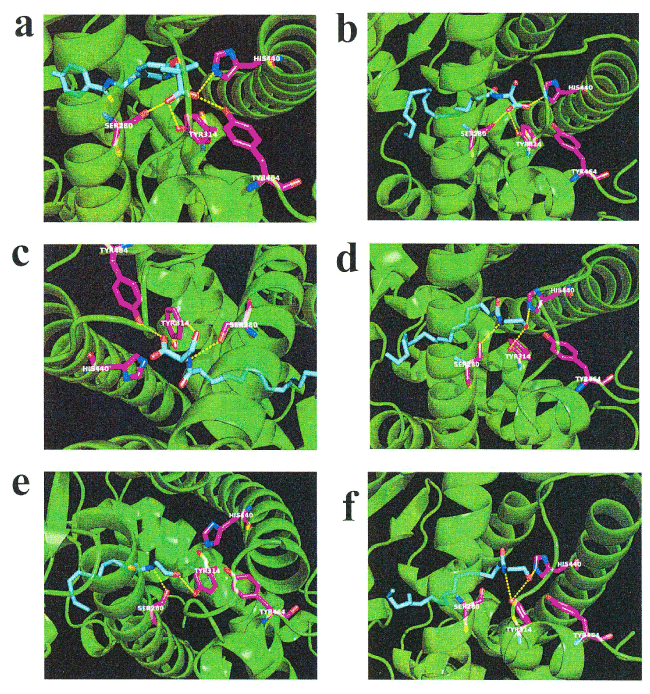
Figure 4. In silico molecular docking of N-18:1 SER, N-16:0 SER, N-18:1 EA, N-16:0 EA, N-18:2 EA, or bezafibrate with PPAR-α. (a) Binding mode of bezafibrate in modeling of PPAR-α; bezafibrate (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The carbonic acid bezafibrate moiety forms a hydrogen bond with the hydroxyl group of Ser-280. The carbonic acid moiety of bezafibrate forms a hydrogen bond with the Tyr-314 hydroxyl group. The carbonic acid bezafibrate moiety forms a hydrogen bond with the His-440 nitrogen atom. The carbonic acid bezafibrate moiety forms a hydrogen bond with the Tyr-464 hydroxyl group. (b) Binding mode of N-18:1 SER in modeling of PPAR- α; N-18:1 SER (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The carbonic acid N-18:1 SER moiety forms a hydrogen bond with the Ser-280 hydroxyl group. The carbonic acid N-18:1 SER moiety forms a hydrogen bond with the Tyr-314 hydroxyl group. The carbonic acid N-18:1 SER moiety forms a hydrogen bond with the His-440 nitrogen atom. (c) Binding mode of N-16:0 SER in modeling of PPAR- α; N-16:0 SER (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The nitrogen atom of N-16:0 SER forms a hydrogen bond with the Ser-280 hydroxyl group. The hydroxyl group of N-16:0 SER forms a hydrogen bond with the Tyr-314 hydroxyl group. The carbonic acid N-16:0 SER moiety forms a hydrogen bond with the Tyr-464 hydroxyl group. (d) Binding mode of N-18:1 EA in modeling of PPAR- α; N-18:1 EA (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The nitrogen atom of N-18:1 EA forms a hydrogen bond with the Ser-280 hydroxyl group. The hydroxyl group at the tip of N-18:1 EA forms a hydrogen bond with the Tyr-314 hydroxyl group. The hydroxyl group at the tip of N-18:1 EA forms a hydrogen bond with the His-440 nitrogen atom. (e) Binding mode of N-16:0 EA in modeling of PPAR- α; N-16:0 EA (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The nitrogen atom of N-16:0 EA forms a hydrogen bond with the Ser-280 hydroxyl group. The hydroxyl group at the tip of N-16:0 EA forms a hydrogen bond with the Tyr-314 hydroxyl group. (f) Binding mode of N-18:2 EA in modeling of PPAR- α; N-18:2 EA (cyan) amino acid (white) in the pocket, hydrogen bonding (yellow dashed line) and hydrogen bonding partner amino acid (magenta). The nitrogen atom of N-18:2 EA forms a hydrogen bond with the Tyr-314 hydroxyl group. The hydroxyl group at the tip of N-18:2 EA forms a hydrogen bond with the Tyr-314 hydroxyl group. The hydroxyl group at the tip of N-18:2 EA forms a hydrogen bond with the His-440 nitrogen atom
Table 2. Docking score and key interacting residues of PPAR-α
Ligand |
Docking score |
Interactive residues |
|
(kcal/mol) |
|
PPAR-α agonist |
Bezafibrate |
-9.323 |
Ser-280, Tyr-314, His-440, and Tyr-464 |
N-Acyl serine |
N-18:1 SER |
-7.366 |
Ser-280, Tyr-314, and His-440 |
N-16:0 SER |
-7.161 |
Ser-280, Tyr-314, and Tyr-464 |
N-Acyl ethanolamine |
N-18:1 EA |
-6.956 |
Ser-280, Tyr-314, and His-440 |
N-16:0 EA |
-6.570 |
Ser-280, and Tyr-314 |
N-18:2 EA |
-6.979 |
Tyr-314, and His-440 |
Effect of N-18:1 SER on FABP1 mRNA expression in human hepatocellular carcinoma HepG2 cells
Because FABP1 and peroxisomal acyl-coenzyme A oxidase 1 (ACOX1) are PPAR-α-responsive gene [12, 31], we next confirmed the effect of N-18:1 SER treatment on FABP1 mRNA using qRT-PCR (Figure 5a). In human HepG2, N-18:1 SER produced a dose-dependent increase in FABP1 mRNA expression (N-18:1 SER-10 µM: 1.48-fold, p=0.0164; N-18:1 SER-30 µM: 1.98-fold, p=0.0112; N-18:1 SER-100 µM: 2.02-fold, p=0.0009). Next, we determined if bezafibrate, a well-known PPAR-α agonist, mimicked the effect of N-18:1 SER in human HepG2 cells. Bezafibrate significantly enhanced FABP1 mRNA expression (bezafibrate-10 µM: 2.86-fold, p=0.0119). We also examined the effect of N-18:1 SER on ACOX1 mRNA expression in human HepG2 cells. N-18:1 SER increased ACOX1 mRNA expression (Figure 5b). We further evaluated whether the PPAR-α antagonist GW6471 attenuated the effect of N-18:1 SER-stimulated FABP1 expression. N-18:1 SER (30 µM)-stimulated FABP1 response was almost completely abolished by 30 µM GW6471 (Figure 5c).
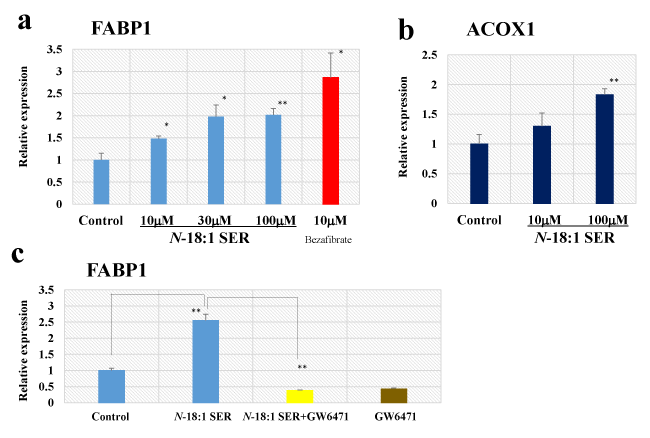
Figure 5. Effect of N-18:1 SER on FABP1 and ACOX1 mRNA expression in human hepatocellular carcinoma HepG2 cells. qRT-PCR analysis of FABP1 (a) and ACOX1 (b) mRNA in human HepG2 cells treated with N-18:1 SER or bezafibrate for 24h. (c) Effect of GW6471 on N-18:1 SER-induced activation of FABP1 mRNA expression in human HepG2 cells. GW6471 (30 µM) was added to cultured cells 30 min prior to and during N-18-1 SER (30 µM) incubation (24h). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was the internal control. Results are presented as mean ± SE (n=5). * p < 0.05 compared with the control (0.5% ethanol). ** p < 0.01 compared with the control (0.5% ethanol)
In this study, we showed that the levels of N-18:1 SER and N-16:0 SER were sensitive to acute EF exposure in healthy humans. The blood concentration of N-18:1 SER in human healthy controls was consistent with that obtained previously by quantitative analysis [32]. In our previous screening using non-targeted metabolomics, we found that EF exposure (9 kV/electrode + 9 kV/electrode)-induced 1.24-fold increase in N-18:1 EA [12]. In the present study, using SRM analysis, EF exposure induced a 1.27-fold increase in the same lipid moiety. Little is known about the biosynthesis of N-acyl SERs, though some possible N-acyl EAs biosynthetic pathways have been proposed [33]. Interestingly, a family of phospholipase A/acyltransferase (PLAAT) that contributes to N-acyl EAs formation has been discovered [34]. However, the detailed mechanisms of EF-induced changes in N-18:1 SER, N-16:0 SER, N-18:1 EA, N-16:0 EA, and N-18:2 EA remain to be elucidated.
The molecular targets of N-18:0 SER and N-16:0 SER are complex and may be interpreted in several ways. In the present study, docking simulation with TRPV1 showed that N-18:1 SER and N-16:0 SER have good binding affinity values. The results indicate that N-18:1 SER and N-16:0 SER binds to the TRPV1 channel. Ahern reported that N-18:1 EA can activate TRPV1 channels following treatment of the protein kinase C activator [35]. Therefore, interaction of the Tyr-511 residue does not have a critical role in the active site of TRPV1. In contrast to N-18:1 EA, the formation of a hydrogen bond with Thr-550 was detected with N-18:1 SER, N-16:0 SER, and the endogenous TRPV1 agonist, 9-HODE [36]. Another study, using cryo-electron microscopy structures of TRPV1 with capsaicin-binding pockets, reported hydrogen bonding to Thr-550 [26-27]. These findings indicate that N-18:1 SER or N-16:0 SER may act as an endogenous agonist for TRPV1. Raboune et al. reported that N-22:6 SER has agonist activity in TRPV1-transfected HEK cells [37]. Evaluation of the agonistic effect of N-18:1 SER or N-16:0 SER in functional assays may be warranted in future studies. The TRPV1 channel is a known pharmacological target to relive pain in post-therapeutic neuralgia [38]. In particular, Anand et al. reported that treatment with Qutenza, a patch containing 8% capsaicin, produced an analgesic effect through defunctionalization of the nociceptor [39]. Borbiro et al. recently reported that activating TRPV1 channels elicits analgesia by inhibiting mechanosensitive Piezo channel activity [40]. Thus, it is reasonable to speculate that EF exposure might alleviate pain through the binding of TRPV1 by N-18:1 SER, or N-16:0 SER. On the other hand, Ito et al. recently reported involvement of TRPV1-mediated calcium signaling in induction of skeletal muscle hypertrophy [41-42]. Although repetitive EF treatment was not performed in the present study, it may be interesting to evaluate the possible effect of HELP exposure on skeletal muscle atrophy in aging.
Eberlein et al. reported that an emollient containing N-16:0 EA improved atopic dermatitis symptoms in a prospective cohort study [43], while Esposito et al. reported that N-16:0 EA treatment improved colon inflammation in an ulcerative colitis model through toll-like receptor 4 (TLR4)-dependent PPAR-α activation [44]. In the present study, the docking simulation with PPAR-α showed that N-acyl SERs have good binding affinity values for N-18:1 SER and N-16:0 SER. In particular, N-18:1 SER binding to PPAR-α was stabilized through the formation of hydrogen bonds with Ser-280, Tyr-314, and His-440. In key interacting residues, a previous study using the PPAR-α model (PDB ID:1i7g) with N-18:1 EA binding pockets reported hydrogen bonding to Ser-280, Tyr-314, and Phe-273 [12]. Because FABP1 and ACOX1 are PPAR-α-responsive genes [12,31], we examined their expression in this study. We previously reported that N-18:1 EA induced 2.8-fold increase in FABP6 gene expression using the Affymetrix GeneChip human genome U133 Plus 2.0 array [12]. Our present study results showed that N-18:1 SER (30 αM) induced an approximately 2-fold increase in FABP1 mRNA expression in human HepG2 cells, which was inhibited by the PPAR-α antagonist GW6471. Thus, HELP exposure may induce FABP1 mRNA expression, at least in part, through activating PPAR-α by N-18:1 SER.
There is evidence that PPAR-α agonists have unique pharmacological effects in vivo. Brandstät et al. reported that fibrates, such as bezafibrate, clofibrate, or fenofibrate, extends the lifespan of adult Caenorhabditis elegans [45]. Zeitler et al. reported that fenofibrate (200 mg/day) induced a quiescent state in patients with multiple benign symmetric lipomatosis (MSL) characterized by a rapid progression [46]. On the other hand, Shirai et al. reported that bezafibrate alleviated circadian rhythm sleep disorders, such as delayed sleep phase syndrome (DSPS) [47]. In bone mass and osteoclast formation, Stunes et al. reported that fenofibrate maintains bone mass in ovariectomized rats [48], while Patel et al. reported that fenofibrate treatment induces decrease of osteoclast formation in cultured derived from mouse marrow [49]. Interestingly, Smoum et al. observed that treatment with N-18:1 SER stimulated the proliferation of osteoblastic MC3T3 E1 cells and primary cavarial osteoblast in vitro [20] and further, N-18:1 SER rescued ovariectomy-induced bone loss in vivo [20]. On the other hand, Hashimoto reported activation of callus formation by EF exposure in a mongrel rabbit in vivo model [50]. In addition, there is evidence that static EF exposure stimulates the differentiation of human cultured osteoblastic cells [51]. Although the underlying mechanisms of bone formation by EF exposure remain to be elucidated, evidence points to N-18:1 SER’s role as an endogenous signaling molecule. Thus, it is conceivable that the increase in N-18:1 SER levels in proportion to the duration of HELP treatment is, at least in part, responsible for slightly high bone density in males aged 70 to 90 years old [52]. In future, the possible effect of EF therapy on MSL, DSPS, and bone mass should be evaluated.
Interestingly, Scuderi et al. reported that N-16:0 EA treatment exerts neuroprotective effects in a rat model of Alzheimer’s disease through PPAR-α involvement [53]. In addition, Campolongo et al. reported that post-training administration of N-18:1 EA in rats enhanced memory consolidation in a Morris water maze performance [54]. These memory-enhancing effects were mimicked by the PPAR-α agonist, GW7647, and was abolished in a mutant animal lacking PPAR-α [54]. Thus, it is reasonable to speculate that EF therapy facilitates memory consolidation via the activation of the PPAR-α signaling pathway by N-18:1 SER, N-16:0 SER, N-16:0 EA, N-18:2 EA, and N-18:1 EA. In future, it will be of interest to evaluate the possible effects of EF therapy on aging-related memory function.
In conclusion, acute HELP exposure induced marked effects on plasma N-18:1 SER and N-16:0 SER levels in healthy subjects. In silico molecular docking of N-18:1 SER and N-16:0 SER was observed in models of TRPV1 and PPAR-α. In human HepG2 cells, N-18:1 SER-stimulated FABP1 mRNA expression was sensitive to the PPAR-α antagonist, GW6471. Our findings provide insight into the molecular mechanisms behind the health benefits induced by HELP.
YN-Y, HH and AH are employees of Hakuju Institute for Health Science Co., Ltd.; TO and HN are employed by Lipidome Lab Co., Ltd.; EK is employed by Intage Healthcare Inc.; and TY is employed by Acel Inc. All other authors have no competing interests.
YN-Y designed and supervised the research, and wrote the manuscript. YN-Y, HH, and AH performed EF exposure. TO and HN performed SRM. EK performed molecular modeling. TY performed RT-PCR. All authors have read and approved the final version of the manuscript.
We thank Dr. Makoto Kikuchi (Professor Emeritus, National Defense Medical College, Japan) for his encouragement.
- Hara H (1961) On the effect of AC. electrostatic high voltage potential load upon the blood-electrolytes (in Japanese). Niigata Medical J 75: 265-273.
- Shiga K, Nakamura K, Kasuga T, Tomoda M, Matsuo T (1967) AC high-voltage electrostatic neuralgia therapy (in Japanese). Iryo 21: 318.
- Nakamura K, Minami S (1970) Effect of high-voltage electrostatic therapy on neuropathic pain in leprosy (in Japanese). Iryo 24: 328-329.
- Ito F, Furuya K (1981) The effect of high voltage alternating current upon a human body the change of blood pressure, endocrine system and serum lipids (in Japanese). J Jpn Sci Balneol Climatol Phys Med 45: 6-17.
- Isaka K, Nishimura R, Arase S, Takiwaki H, Osaki K, et al. (1998) Dosimetry and exposure experiments for extremely low frequency high-tension electric field therapy. EMC ’98 Rome International Symposium on Electromagnetic Comoatibility D: 204-207.
- Nawarat S, Iomsai K, Jantanam P, Kauengtip Y (1999) Effects of electrical Healthtron on curing of non-communicable diseases: Case study of Banlad hospital Petchaburi province (in Thai). Region 4 Medical J 18: 139-149.
- Ito F (2000) The role of electric field therapeutic device (Healthtron) in the therapy of acute low back pain (in Japanese). J Jpn Sci Balneol Climatol Phys Med 63: 127-137.
- Siripanichgon K, Otrakul A, Suparp J, Sirikulchayanonta C, Charupoonphol P (2000) Clinical observation of Healthtron therapy (in Thai). J Public Health (Bangkok) 30: 19-29.
- Sirikulchayanonta C, Siripanichgon K, Otrakul A, Suparp J, Charupoonphol P (2001) The effect of Healthtron on serum lipid levels among the middle-aged: Preliminary report. J Public Health (Bangkok) 31: 63-70.
- Ito F, Ohsaki K, Takahashi K, Hara H (2005) The effects of electric field therapeutic device (Healthtron) on the stiffness in the neck and shoulder area – changes in subjective symptoms, blood circulation and the autonomic nervous system (in Japanese). J Jpn Sci Balneol Climatol Phys Med 68: 110-121.
- Shinba T, Takahashi K, Kanetaka S, Nedachi T, Yamaneki M, et al. (2012) A pilot study on electric field therapy for chronic pain with no obvious underlying diseases (in Japanese). Soc Integrative Med Jpn 5: 68-72.
- Nakagawa-Yagi Y, Hara H, Fujimori T, Yamaguchi T, Midorikawa A, et al. (2014) Non-targeted human plasma metabolomics reveals the changes in oleoylethanolamide, a lipid-derived signaling molecule, by acute exposure of electric field. Integr Mol Med 1: 29-37.
- Nakagawa-Yagi Y, Hara H, Yoshida Y, Midorikawa A, Hara A (2015) Discovery of a novel effect of electric field exposure on human plasma beta-endorphin and interleukin-12 levels: Insight into mechanisms of pain alleviation and defense against infection by electric field therapy. Integr Mol Med 2: 200-204.
- Nakagawa-Yagi Y, Hara H, Nakagawa F, Sato M, Hara A (2016) Acute exposure to an electric field induces changes in human plasma 9-HODE, 13-HODE, and immunoreactive substance P levels: Insight into the molecular mechanisms of electric field therapy. Integr Mol Med 3: 600-605.
- Nakagawa-Yagi Y, Hara H, Tsuboi H, Abe J, Hara A (2016) Effect of 3-hydroxybutyrate, an endogenous histone deacetylase inhibitor, on FOXO3A mRNA expression in human epithelial colorectal Caco-2 cells: Insight into the epigenetic mechanisms of electric field therapy. Integr Mol Med 3: 764-768.
- Ohtsuki T, Nabeta T, Nakanishi H, Kawahata H, Ogihara T, et al. (2017) Electric field exposure improves subjective symptoms related to sleeplessness in college students: A pilot study of electric field therapy for sleep disorder. Curr Med Chem 17: 37-48.
- Nakagawa-Yagi Y, Hara H, Nakanishi H, Tasaka T, Hara A (2017) Acute exposure to an electric field induces changes in human plasma lysophosphatidylcholine (lysoPC)-22:4 levels: Molecular insight into the docking of lysoPC-22:4 interaction with TRPV2. Integr Mol Med 4: 1-7.
- Nakagawa-Yagi Y, Hara H, Nakanishi H, Kanai C, Hara A (2017) Molecular insight into the docking of lysophosphatidylethanolamine (lysoPE)-22:6 interaction with GPR119: Acute exposure to an electric field induces changes in human plasma lysoPE-22:6 and lysoPE-20:4 levels. Integr Mol Med 4: 1-7.
- Nakagawa-Yagi Y, Hara H, Kanai C, Sato M, Hara A (2018): Acute electric field downregulates human plasma immunoreactive interleukin-6 and -1β levels: Molecular mechanisms underlying inflammation alleviation through electric field therapy. Integr Mol Med 5: 1-6.
- Smoum R, Bar A, Tan B, Milman G, Attar-Namdar M, et al. (2010) Oleoyl serine, an endogenous N-acyl amide, modulates bone remodeling and mass. Proc Natl Acad Sci U S A 107: 17710-17715. [Crossref]
- Marczylo TH, Lam PM, Nallendran V, Taylor AH, Konje JC (2009) A solid-phase method for the extraction and measurement of anandamide from multiple human biomatrices. Anal Biochem 384: 106-113.
- Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31: 455-461.
- Kuwabara N, Oyama T, Tomioka D, Ohashi M, Yanagisawa J, et al. (2012) Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype. J Med Chem 55: 893-902. [Crossref]
- Movahed P, Jonsson BG, Birnir B, Wingstrand JA, Jorgensen TD, et al. (2005) Endogenous unsaturated C18 N-acylethanolamines are vanilloid receptor (TRPV1) agonists. J Biol Chem 280: 38496-38504.
- Tominaga M, Caterina MJ, Malmberg AB, Rosen TA, Gilbert H, et al. (1998) The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21: 531-543.
- Yang F, Zheng J (2017) Understand spiciness: mechanism of TRPV1 channel activation by capsaicin. Protein Cell 8: 169-177. [Crossref]
- Yang F, Xiao X, Cheng W, Yang W, Yu P, et al. (2015) Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nature Chem Biol 11: 518-524.
- Fu J, Gaetani S, Oveisi F, Lo Verme J, Serrano A, et al. (2003) Oleoylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-α. Nature 425: 90-93.
- Guzman M, Lo Verme J, Fu J, Oveisi F, Blazquez C, et al. (2004) Oleoylethanolamide stimulates lipolysis by activating the nuclear receptor peroxisome proliferator-activated receptor alpha (PPAR-alpha). J Biol Chem 279: 27849-27854.
- Tenenbaum A, Fishman EZ (2012) Balanced pan-PPAR activator bezafibrate in combination with statin: comprehensive lipids control and diabetes prevention? Cardiovasc Diabetol 11: 140
- Landrier JF, Thomas C, Grober J, Duez H, Percevault F, et al. (2004) Statin induction of liver fatty acid-binding protein (L-FABP) gene expression is peroxisome proliferator-activated receptor-alpha-dependent. J Biol Chem 279: 45512-45518. [Crossref]
- Braghithy S, Smoum R, Drori A, Hadar R, Gammal A, et al. (2019) Magel2 modulates bone remodeling and mass in Prader-Willi syndrome by affecting oleoyl serine levels and activity. J Bone Miner Res 34: 93-105.
- Ogura Y, Parsons WH, Kamat SS, Cravatt BF (2016) A calcium-dependent acyltransferase that produces N-acylphosphatidylethanolamines. Nat Chem Biol 12: 669-671.
- Uyama T, Tsuboi K, Ueda N (2017) An involvement of phospholipase A/acyltransferase family proteins in peroxisome regulation and plasmalogen metabolism. FEBS Lett 591: 2745-2760. [Crossref]
- Ahern GP (2003) Activation of TRPV1 by the satiety factor oleoylethanolamide. J Biol Chem 278: 30429-30434. [Crossref]
- Patwardhan AM, Akopian AN, Ruparel NB, Diogenes A, Weintraub ST, et al. (2010) Heat generates oxidized linoleic acid metabolites that activate TRPV1 and produce pain in rodents. J Clin Invest 120: 1617-1626. [Crossref]
- Raboune S, Stuart JM, Leishman E, Takacs SM, Rhodes B, et al. (2014) Novel endogenous N-acyl amides activate TRPV1-4 receptors, BV-2 microglia, and are regulated in brain in an acute model of inflammation. Front Cell Neurosci 8: 195.
- Vay L, Gu C, McNaughton PA (2012) The thermo-TRP ion channel family: properties and therapeutic implications. Br J Pharmacol 165: 787-801. [Crossref]
- Anand P, Bley K (2011) Topical capsaicin for pain management: therapeutic potential and mechanisms of action of the new high-concentration capsaicin 8% patch. Br J Anaesth 107: 490-502. [Crossref]
- Borbiro I, Badheka D, Rohacs T (2015) Activation of TRPV1 channels inhibits mechanosensitive Piezo channel activity by depleting membrane phosphoinositides. Sci Signal 8: 15.
- Ito N, Ruegg UT, Kudo A, Miyagoe-Suzuki Y, Takeda S (2013) Activation of calcium signaling through Trpv1 by nNOS and peroxynitrite as a key trigger of skeletal muscle hypertrophy. Nat Med 19: 101-106.
- Ito N, Ruegg UT, Kudo A, Miyagoe-Suzuki Y, Takeda S (2013) Capsaicin mimics mechanical load-induced intracellular signaling events: Involvement of TRPV1-mediated calcium signaling in induction of skeletal muscle hypertrophy. Channels 7: 221-224.
- Eberlein B, Eicke C, Reinhardt HW, Ring J (2008) Adjuvant treatment of atopic eczema: assessment of an emollient containing N-palmitoylethanolamine (ATOPA study). J Eur Acad Dermatol Venereol 22: 73-82. [Crossref]
- Esposito G, Capoccia E, Turco F, Palumbo I, Lu J, et al. (2014) Palmitoylethanolamide improves colon inflammation through an enteric glia/toll like receptor 4-dependent PPAR-α activation. Gut 63: 1300-1312.
- Brandstät S, Schmeisser K, Zarse K, Ristow M (2013) Lipid-lowering fibrates extend C. elegans lifespan in a NHR-49/PPARalpha-dependent manner. Aging 5: 270-275
- Zeitler H, Ulrich-Merzenich G, Richter DF, Vetter H, Walger P (2008) Multiple benign symmetric lipomatosis – a differential diagnosis of obesity. Is there a rationale for fibrate treatment? Obes Surg 18: 1354-1356
- Shirai H, Oishi K, Kudo T, Shibata S, Ishida N (2007) PPAR α is a potential therapeutic target of drugs to treat circadian rhythm sleep disorder. Biochem Biophys Res Commun 357: 679-682.
- Stunes AK, Westbroek I, Gustafsson BI, Fossmark R, Waarsing JH, et al. (2011) The peroxisome proliferator-activated receptor (PPAR) alpha agonist fenofibrate maintains bone mass, while the PPAR gamma agonist pioglitazone exaggerates bone loss, in ovariectomized rats. BMC Endocr Disord 11: 11. [Crossref]
- Patel JJ, Butters OR, Arnett TR (2014) PPAR agonists stimulate adipogenesis at the expense of osteoblast differentiation while inhibiting osteoclast formation and activity. Cell Biochem Funct 32: 368-377.
- Hashimoto T (1975) The effect of static electric field caused by high voltage alternating current on callus formation (in Japanese). J Kyoto Pref Univ Med 84: 89-108.
- Su CY, Fang T, Fang HW (2017) Effects of Electrostatic Field on Osteoblast Cells for Bone Regeneration Applications. Biomed Res Int 2017: 7124817. [Crossref]
- Harakawa S, Hori T, Inoue N, Okano H, Nedachi T, et al. (2014) Effect of extensive electric field therapy in bone density (in Japanese). Soc Integrative Med Jan 7: 60-66.
- Scuderi C, Stecca C, Valenza M, Patano P, Bronzuoli MR, et al. (2014) Palmitoylethanolamide controls reactive gliosis and exerts neuroprotective functions in a rat model of Alzheimer’s disease. Cell Death Dis 5: e1419.
- Campolongo P, Roozendaal B, Trezza V, Cuomo V, Astarita G, et al. (2009) Fat-induced satiety factor oleoylethanolamide enhances memory consolidation. Proc Natl Acad Sci USA 106: 8027-8031.





